Replication stress generates distinctive landscapes of DNA copy number alterations and chromosome scale losses
- PMID: 36266663
- PMCID: PMC9583511
- DOI: 10.1186/s13059-022-02781-0
Replication stress generates distinctive landscapes of DNA copy number alterations and chromosome scale losses
Abstract
Background: A major driver of cancer chromosomal instability is replication stress, the slowing or stalling of DNA replication. How replication stress and genomic instability are connected is not known. Aphidicolin-induced replication stress induces breakages at common fragile sites, but the exact causes of fragility are debated, and acute genomic consequences of replication stress are not fully explored.
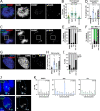
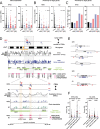
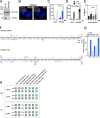
Results: We characterize DNA copy number alterations (CNAs) in single, diploid non-transformed cells, caused by one cell cycle in the presence of either aphidicolin or hydroxyurea. Multiple types of CNAs are generated, associated with different genomic regions and features, and observed copy number landscapes are distinct between aphidicolin and hydroxyurea-induced replication stress. Coupling cell type-specific analysis of CNAs to gene expression and single-cell replication timing analyses pinpointed the causative large genes of the most recurrent chromosome-scale CNAs in aphidicolin. These are clustered on chromosome 7 in RPE1 epithelial cells but chromosome 1 in BJ fibroblasts. Chromosome arm level CNAs also generate acentric lagging chromatin and micronuclei containing these chromosomes.
Conclusions: Chromosomal instability driven by replication stress occurs via focal CNAs and chromosome arm scale changes, with the latter confined to a very small subset of chromosome regions, potentially heavily skewing cancer genome evolution. Different inducers of replication stress lead to distinctive CNA landscapes providing the opportunity to derive copy number signatures of specific replication stress mechanisms. Single-cell CNA analysis thus reveals the impact of replication stress on the genome, providing insights into the molecular mechanisms which fuel chromosomal instability in cancer.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

