Crosstalk between metabolic reprogramming and epigenetics in cancer: updates on mechanisms and therapeutic opportunities
- PMID: 36266736
- PMCID: PMC9648395
- DOI: 10.1002/cac2.12374
Crosstalk between metabolic reprogramming and epigenetics in cancer: updates on mechanisms and therapeutic opportunities
Abstract
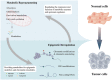
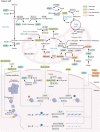
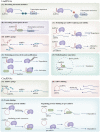
Reversible, spatial, and temporal regulation of metabolic reprogramming and epigenetic homeostasis are prominent hallmarks of carcinogenesis. Cancer cells reprogram their metabolism to meet the high bioenergetic and biosynthetic demands for vigorous proliferation. Epigenetic dysregulation is a common feature of human cancers, which contributes to tumorigenesis and maintenance of the malignant phenotypes by regulating gene expression. The epigenome is sensitive to metabolic changes. Metabolism produces various metabolites that are substrates, cofactors, or inhibitors of epigenetic enzymes. Alterations in metabolic pathways and fluctuations in intermediate metabolites convey information regarding the intracellular metabolic status into the nucleus by modulating the activity of epigenetic enzymes and thus remodeling the epigenetic landscape, inducing transcriptional responses to heterogeneous metabolic requirements. Cancer metabolism is regulated by epigenetic machinery at both transcriptional and post-transcriptional levels. Epigenetic modifiers, chromatin remodelers and non-coding RNAs are integral contributors to the regulatory networks involved in cancer metabolism, facilitating malignant transformation. However, the significance of the close connection between metabolism and epigenetics in the context of cancer has not been fully deciphered. Thus, it will be constructive to summarize and update the emerging new evidence supporting this bidirectional crosstalk and deeply assess how the crosstalk between metabolic reprogramming and epigenetic abnormalities could be exploited to optimize treatment paradigms and establish new therapeutic options. In this review, we summarize the central mechanisms by which epigenetics and metabolism reciprocally modulate each other in cancer and elaborate upon and update the major contributions of the interplays between epigenetic aberrations and metabolic rewiring to cancer initiation and development. Finally, we highlight the potential therapeutic opportunities for hematological malignancies and solid tumors by targeting this epigenetic-metabolic circuit. In summary, we endeavored to depict the current understanding of the coordination between these fundamental abnormalities more comprehensively and provide new perspectives for utilizing metabolic and epigenetic targets for cancer treatment.
Keywords: RNA epigenetics; cancer; epigenetics; metabolic reprogramming; therapy.
© 2022 The Authors. Cancer Communications published by John Wiley & Sons Australia, Ltd. on behalf of Sun Yat-sen University Cancer Center.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646‐74. - PubMed
-
- Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022;12(1):31‐46. - PubMed
-
- Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, et al. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22(1):66‐79. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

