The cuproptosis-related gene signature serves as a potential prognostic predictor for ovarian cancer using bioinformatics analysis
- PMID: 36267774
- PMCID: PMC9577750
- DOI: 10.21037/atm-22-4546
The cuproptosis-related gene signature serves as a potential prognostic predictor for ovarian cancer using bioinformatics analysis
Abstract
Background: Studies have shown that copper is involved in the tumorigenesis and development of ovarian cancer. In this work, we aimed to build a prognostic classification system associated with cuproptosis to predict ovarian cancer prognosis.
Methods: Information of ovarian cancer samples were acquired from The Cancer Genome Atlas (TCGA)-ovarian cancer and GSE26193 dataset. Cuproptosis-related genes were screened from previous research. ConsensusClusterPlus was applied to determine molecular subtypes, which were evaluated by tumor immune microenvironment analysis, TIDE algorithm, and functional enrichment analysis. Furthermore, limma analysis and univariate Cox analysis were used to construct a cuproptosis-related prognostic signature for ovarian cancer. Univariate and multivariate Cox regression analyses were used to analyze the independence of clinical factors and model.
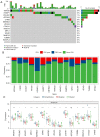
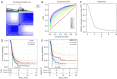
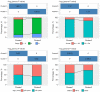
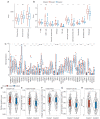
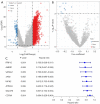
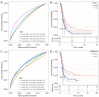
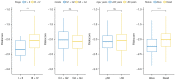
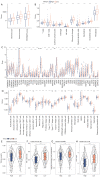
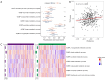
Results: A total of 15 genes related to cuproptosis were identified, and 2 clusters (C1 and C2) were determined. C1 had a better survival outcome, less advanced stage, enhanced immune infiltration, was more sensitive to immunotherapy, and showed enrichment in tricarboxylic acid (TCA)-related pathways. An 8 cuproptosis-associated gene signature was constructed, and the signature was verified in the GSE26193 dataset. A higher risk score of the cuproptosis-related gene signature was significantly correlated with worse overall survival (OS) (P<0.0001), which was validated in GSE26193 dataset successfully. Cox survival analysis showed that risk score was an independent predictor [hazard ratio (HR) =2.66, P<0.001]. Functional enrichment and tumor immune microenvironment analyses showed that high-risk patients tended to have immunologically sensitive tumors.
Conclusions: The cuproptosis-related gene signature may serve as a potential prognostic predictor for ovarian cancer patients and may offer novel treatment strategies for ovarian cancer.
Keywords: Ovarian cancer; clusters; cuproptosis; prognosis; signature.
2022 Annals of Translational Medicine. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-4546/coif). The authors have no conflicts of interest to declare.
Figures

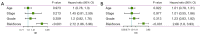
References
-
- Moss HA, Berchuck A, Neely ML, et al. Estimating Cost-effectiveness of a Multimodal Ovarian Cancer Screening Program in the United States: Secondary Analysis of the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS). JAMA Oncol 2018;4:190-5. 10.1001/jamaoncol.2017.4211 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
