Comparative transcriptome analysis of Citrus macrophylla tree infected with Citrus tristeza virus stem pitting mutants provides new insight into the role of phloem regeneration in stem pitting disease
- PMID: 36267951
- PMCID: PMC9577373
- DOI: 10.3389/fpls.2022.987831
Comparative transcriptome analysis of Citrus macrophylla tree infected with Citrus tristeza virus stem pitting mutants provides new insight into the role of phloem regeneration in stem pitting disease
Abstract
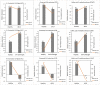
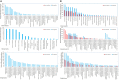
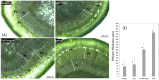
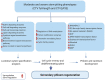
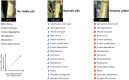
Stem pitting is a complex and economically important virus-associated disease of perennial woody plants. Molecular mechanisms and pathways occurring during virus-plant interaction that result in this phenomenon are still obscure. Previous studies indicated that different Citrus tristeza virus (CTV) mutants induce defined stem pitting phenotypes ranging from mild (CTVΔp13) to severe (CTVΔp33) in Citrus macrophylla trees. In this study, we conducted comparative transcriptome analyses of C. macrophylla trees infected with CTV mutants (CTVΔp13 and CTVΔp33) and a full-length virus in comparison to healthy plants as control. The mild CTV stem pitting mutant had very few differentially expressed genes (DEGs) related to plant defense mechanism and plant growth and development. In contrast, substantial gene expression changes were observed in plants infected with the severe mutant and the full-length virus, indicating that both the p13 and p33 proteins of CTV acted as a regulator of symptom production by activating and modulating plant responses, respectively. The analysis of transcriptome data for CTVΔp33 and the full-length virus suggested that xylem specification has been blocked by detecting several genes encoding xylem, cell wall and lignin degradation, and cell wall loosening enzymes. Furthermore, stem pitting was accompanied by downregulation of transcription factors involved in regulation of xylem differentiation and downregulation of some genes involved in lignin biosynthesis, showing that the xylem differentiation and specification program has been shut off. Upregulation of genes encoding transcription factors associated with phloem and cambium development indicated the activation of this program in stem pitting disease. Furthermore, we detected the induction of several DEGs encoding proteins associated with cell cycle re-entry such as chromatin remodeling factors and cyclin, and histone modification. This kind of expression pattern of genes related to xylem differentiation and specification, phloem and cambium development, and cell cycle re-entry is demonstrated during secondary vascular tissue (SVT) regeneration. The microscopy analysis confirmed that the regeneration of new phloem is associated with stem pitting phenotypes. The findings of this study, thus, provide evidence for the association between stem pitting phenotypes and SVT regeneration, suggesting that the expression of these genes might play important roles in development of stem pitting symptoms. Overall, our findings suggest that phloem regeneration contributes to development of stem pitting symptoms.
Keywords: Citrus tristeza virus (CTV); secondary vascular tissue regeneration; stem-pitting; transcriptome (RNA-seq); virus-plant interaction.
Copyright © 2022 Khalilzadeh, Weber, Dutt, El-Mohtar and Levy.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Amenduni T., Hobeika C., Minafra A., Boscia D., Castellano M. A., Savino V. (2005). Plum bark necrosis stem pitting-associated virus in different stone fruit species in Italy. J. Plant Pathol. 87, 131–134. 10.17660/ActaHortic.2004.657.10 - DOI
-
- Bar-Joseph M., Marcus R., Lee R. F. (1989). The continuous challenge of Citrus tristeza virus control. Annu. Rev. Phytopathol. 27, 291–316. 10.1146/annurev.py.27.090189.001451 - DOI
LinkOut - more resources
Full Text Sources

