A new family of CRISPR-type V nucleases with C-rich PAM recognition
- PMID: 36268581
- PMCID: PMC9724661
- DOI: 10.15252/embr.202255481
A new family of CRISPR-type V nucleases with C-rich PAM recognition
Abstract
Most CRISPR-type V nucleases are stimulated to cleave double-stranded (ds) DNA targets by a T-rich PAM, which restricts their targeting range. Here, we identify and characterize a new family of type V RNA-guided nuclease, Cas12l, that exclusively recognizes a C-rich (5'-CCY-3') PAM. The organization of genes within its CRISPR locus is similar to type II-B CRISPR-Cas9 systems, but both sequence analysis and functional studies establish it as a new family of type V effector. Biochemical experiments show that Cas12l nucleases function optimally between 37 and 52°C, depending on the ortholog, and preferentially cut supercoiled DNA. Like other type V nucleases, it exhibits collateral nonspecific ssDNA and ssRNA cleavage activity that is triggered by ssDNA or dsDNA target recognition. Finally, we show that one family member, Asp2Cas12l, functions in a heterologous cellular environment, altogether, suggesting that this new group of CRISPR-associated nucleases may be harnessed as genome editing reagents.
Keywords: CRISPR-Cas; PAM; RNA-guided nuclease; genome editing; nucleic acid detection.
© 2022 The Authors. Published under the terms of the CC BY 4.0 license.
Figures
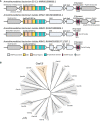
A schematic of Cas12l locus architecture. CRISPR‐associated genes are shown in light blue and each locus identified encodes one approximately 860 aa effector containing a single tri‐split RuvC nuclease domain and a helix‐turn‐helix motif (orange) followed by Cas1, Cas2, and Cas4.
Maximum likelihood phylogenetic tree illustrating the sequence relationship between Cas12l and other Type V CRISPR‐Cas proteins.

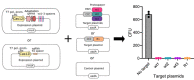
Workflow used to detect dsDNA cleavage and associated PAM recognition of Cas12l CRISPR systems. E. coli was transformed with plasmids encoding an intact CRISPR locus encoding the Cas12l effector nuclease and adaptation proteins, Cas1, Cas2, and Cas4, engineered to target a randomized PAM plasmid library (T1 and T2 spacers correspond to Target1 and Target2). After expression, cells were disrupted and the resulting lysate used in subsequent steps. Cleavage products were captured by adapter ligation, enriched for using PCR, and subjected to Illumina deep sequencing.
dsDNA cleavage was detected by examining the frequency of adapter ligation at each position of the protospacer target relative to control reactions assembled with an empty vector. Once identified, reads associated with the highest frequency of adapter ligation were used to assess PAM recognition. Weblogos of the PAM sequences that supported target recognition and cleavage are shown.

Alignment of the sequences between the genes encoding the Cas12l effector and Cas1 exhibits features of a trans‐activating CRISPR RNA (tracrRNA). Percent identity is shown in teal. These include a sequence encoding a 5′ hairpin (Hairpin 1—gray), a conserved sequence encoding a nexus‐like stem‐loop (Hairpin 2—blue), a region capable of base pairing with the CRISPR repeat (Anti‐repeat—orange), and a GC‐rich sequence capable of forming a terminator‐like hairpin (Hairpin 3—red).
Four single‐guide RNA (sgRNA) designs were engineered for Asp2Cas12l and Asp3Cas12l based on the secondary structure prediction of tracrRNAs in (A). They differed by the presence or omission of a 5′ region of the tracrRNA predicted to be unstructured and by the inclusion or exclusion of the terminator‐like Hairpin 3. tracrRNA features are colored as described in (A).
Cleavage of supercoiled (SC) plasmid DNA substrates using purified Cas12l nuclease and respective sgRNA variants. Efficient full‐length linearization (FLL) of the substrate resulting from a complete double‐strand break was only observed when using sgRNAs bearing the 5′ most end of the tracrRNA (variants 1 and 3). Moreover, the terminator‐like Hairpin 3 is not required for target cleavage (variant 1). OC, open circular; FLL, full‐length linear; SC, supercoiled.
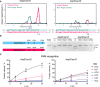
Positions of Asp2Cas12l and Asp3Cas12l target and nontarget strand cleavage. Frequency of adater‐ligated reads from PAM library cleavage experiments for both target and nontarget strands. The target strand is cleaved 23–24 nt 3′ of the PAM and the nontarget strand is cleaved 15–18 nt downstream of PAM recognition.
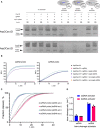
Substitution of alanine residues disrupts linear dsDNA cleavage confirming key catalytic positions within the RuvC nuclease domain of Asp2Cas12l and Asp3Cas12l. wt, wildtype.
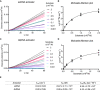
Cleavage of oligoduplex dsDNA substrates with purified RNP complexes confirms PAM recognition. The molar ratio of RNP to the substrate was kept low (5:1) to increase reaction stringency. Asp2Cas12l predominantly recognizes a 5'‐CCY‐3' PAM and Asp3Cas12l a 5'‐CCB‐3' PAM. N = 3.
Asp2 and Asp3Cas12l RNP complexes degrade M13 ssDNA in the presence of single‐stranded (ss) DNA or dsDNA (including PAM) activators with target sequences complementary to the gRNA spacer. NS activator—nonspecific activator (ssDNA oligonucleotide or dsDNA duplex) with no sequence complementarity to the spacer of the gRNA.
Asp2Cas12l RNP complexes activated with ss or dsDNA degrade quenched fluorescent ssDNA or ssRNA probes. Background‐subtracted traces from fluorescent reporter assays with favored ssDNA (5'‐CCCCCCCC‐3′) or ssRNA (5'‐CCCCCCCC‐3′) probes. RFU, relative fluorescence units.
Background‐subtracted traces of fluorescent reporter cleavage using ssDNA or ssRNA probes.
Rates of trans‐degradation with ssRNA or ssDNA reporters activated with either ssDNA or dsDNA targets. Collateral ssDNAse activity is about 3‐fold higher than the rate of ssRNA degradation.
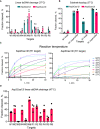
Cas12l dsDNA hydrolysis efficiency varies depending on the target sequence.
Linear and supercoiled dsDNA substrates with different protospacer sequences (W1 and R1) were interrogated by Asp2Cas12l and Asp3Cas12l RNPs. With a linear topology, only the R1 protospacer target was appreciably cleaved; however, both protospacers were cleaved with similar efficiencies by both proteins when presented in a supercoiled state.
Effect of reaction temperature on Asp2Cas12l and Asp3Cas12l dsDNA hydrolysis. Optimal temperature for dsDNA cleavage by Asp2Cas12l and Asp3Cas12l RNP complexes is ~50°C.
Asp3Cas12l dsDNA hydrolysis across various targets at the increased reaction temperature.
Fluorescently labeled (5′‐6‐FAM depicted in blue and 5′‐6‐ROX in red) linear oligoduplex dsDNA substrate with 5'‐CCC‐3' PAM (underlined) and R1 protospacer sequence (bold) used for hydrolysis experiments and cleavage rates of nontarget (NTS) and target (TS) DNA strands.
Fluorescently labeled (5′‐6‐FAM depicted in blue and 5′‐6‐ROX in red) linear oligoduplex dsDNA substrate with 5'‐CCC‐3' PAM (underlined) and W1 protospacer sequence (bold) used for hydrolysis experiments and cleavage rates of nontarget (NTS) and target (TS) DNA strands.
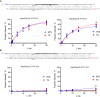
Background‐subtracted traces and corresponding linear trendlines of cleaved substrate concentration versus time for an ssDNA activator, using 0.1 nM effective Asp2Cas12l‐sgRNA‐activator complex and increasing ssDNA reporter concentration.
Michaelis–Menten fits for the ssDNA activator.
Background‐subtracted traces and corresponding linear trendlines of cleaved substrate concentration versus time for a dsDNA activator, using 0.1 nM effective Asp2Cas12l‐sgRNA‐activator complex and increasing ssDNA reporter concentration.
Michaelis–Menten fits for the dsDNA activator.
Calculated kinetic constant values.
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 - PubMed
-
- Anzalone AV, Koblan LW, Liu DR (2020) Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol 38: 824–844 - PubMed
-
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712 - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

