APC/CCdc20-mediated degradation of Clb4 prompts astral microtubule stabilization at anaphase onset
- PMID: 36269172
- PMCID: PMC9595209
- DOI: 10.1083/jcb.202203089
APC/CCdc20-mediated degradation of Clb4 prompts astral microtubule stabilization at anaphase onset
Abstract
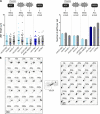
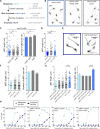
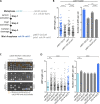
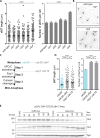
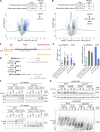
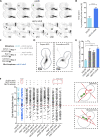
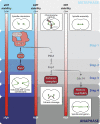
Key for accurate chromosome partitioning to the offspring is the ability of mitotic spindle microtubules to respond to different molecular signals and remodel their dynamics accordingly. Spindle microtubules are conventionally divided into three classes: kinetochore, interpolar, and astral microtubules (kMTs, iMTs, and aMTs, respectively). Among all, aMT regulation remains elusive. Here, we show that aMT dynamics are tightly regulated. aMTs remain unstable up to metaphase and are stabilized at anaphase onset. This switch in aMT dynamics, important for proper spindle orientation, specifically requires the degradation of the mitotic cyclin Clb4 by the Anaphase Promoting Complex bound to its activator subunit Cdc20 (APC/CCdc20). These data highlight a unique role for mitotic cyclin Clb4 in controlling aMT regulating factors, of which Kip2 is a prime candidate, provide a framework to understand aMT regulation in vertebrates, and uncover mechanistic principles of how the APC/CCdc20 choreographs the timing of late mitotic events by sequentially impacting on the three classes of spindle microtubules.
© 2022 Zucca et al.
Figures









References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

