A tug of war between filament treadmilling and myosin induced contractility generates actin rings
- PMID: 36269229
- PMCID: PMC9678364
- DOI: 10.7554/eLife.82658
A tug of war between filament treadmilling and myosin induced contractility generates actin rings
Abstract
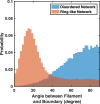
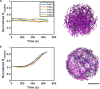
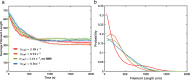
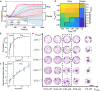
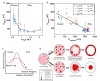
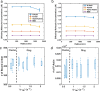
In most eukaryotic cells, actin filaments assemble into a shell-like actin cortex under the plasma membrane, controlling cellular morphology, mechanics, and signaling. The actin cortex is highly polymorphic, adopting diverse forms such as the ring-like structures found in podosomes, axonal rings, and immune synapses. The biophysical principles that underlie the formation of actin rings and cortices remain unknown. Using a molecular simulation platform called MEDYAN, we discovered that varying the filament treadmilling rate and myosin concentration induces a finite size phase transition in actomyosin network structures. We found that actomyosin networks condense into clusters at low treadmilling rates or high myosin concentrations but form ring-like or cortex-like structures at high treadmilling rates and low myosin concentrations. This mechanism is supported by our corroborating experiments on live T cells, which exhibit ring-like actin networks upon activation by stimulatory antibody. Upon disruption of filament treadmilling or enhancement of myosin activity, the pre-existing actin rings are disrupted into actin clusters or collapse towards the network center respectively. Our analyses suggest that the ring-like actin structure is a preferred state of low mechanical energy, which is, importantly, only reachable at sufficiently high treadmilling rates.
Keywords: T cells; actomyosin ring; computational biology; cytoskeleton; molecular simulation; none; systems biology.
© 2022, Ni et al.
Conflict of interest statement
QN, KW, AP, HN, VV, AU, GP No competing interests declared
Figures









References
-
- Bovellan M, Romeo Y, Biro M, Boden A, Chugh P, Yonis A, Vaghela M, Fritzsche M, Moulding D, Thorogate R, Jégou A, Thrasher AJ, Romet-Lemonne G, Roux PP, Paluch EK, Charras G. Cellular control of cortical actin nucleation. Current Biology. 2014;24:1628–1635. doi: 10.1016/j.cub.2014.05.069. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

