Stochastic dynamics of Type-I interferon responses
- PMID: 36269758
- PMCID: PMC9629604
- DOI: 10.1371/journal.pcbi.1010623
Stochastic dynamics of Type-I interferon responses
Abstract
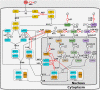
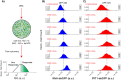
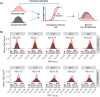
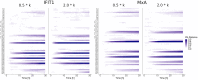
Interferon (IFN) activates the transcription of several hundred of IFN stimulated genes (ISGs) that constitute a highly effective antiviral defense program. Cell-to-cell variability in the induction of ISGs is well documented, but its source and effects are not completely understood. The molecular mechanisms behind this heterogeneity have been related to randomness in molecular events taking place during the JAK-STAT signaling pathway. Here, we study the sources of variability in the induction of the IFN-alpha response by using MxA and IFIT1 activation as read-out. To this end, we integrate time-resolved flow cytometry data and stochastic modeling of the JAK-STAT signaling pathway. The complexity of the IFN response was matched by fitting probability distributions to time-course flow cytometry snapshots. Both, experimental data and simulations confirmed that the MxA and IFIT1 induction circuits generate graded responses rather than all-or-none responses. Subsequently, we quantify the size of the intrinsic variability at different steps in the pathway. We found that stochastic effects are transiently strong during the ligand-receptor activation steps and the formation of the ISGF3 complex, but negligible for the final induction of the studied ISGs. We conclude that the JAK-STAT signaling pathway is a robust biological circuit that efficiently transmits information under stochastic environments.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Maiwald T, Schneider A, Busch H, Sahle S, Gretz N, Weiss TS, et al.. Combining theoretical analysis and experimental data generation reveals IRF9 as a crucial factor for accelerating interferon α-induced early antiviral signalling. FEBS journal. 2010;277(22):4741–4754. doi: 10.1111/j.1742-4658.2010.07880.x - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

