The ZFP36 family of RNA binding proteins regulates homeostatic and autoreactive T cell responses
- PMID: 36269839
- PMCID: PMC9832469
- DOI: 10.1126/sciimmunol.abo0981
The ZFP36 family of RNA binding proteins regulates homeostatic and autoreactive T cell responses
Abstract
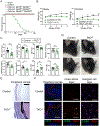
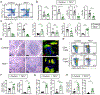
RNA binding proteins are important regulators of T cell activation, proliferation, and cytokine production. The zinc finger protein 36 (ZFP36) family genes (Zfp36, Zfp36l1, and Zfp36l2) encode RNA binding proteins that promote the degradation of transcripts containing AU-rich elements. Numerous studies have demonstrated both individual and shared functions of the ZFP36 family in immune cells, but their collective function in T cells remains unclear. Here, we found a redundant and critical role for the ZFP36 proteins in regulating T cell quiescence. T cell-specific deletion of all three ZFP36 family members in mice resulted in early lethality, immune cell activation, and multiorgan pathology characterized by inflammation of the eyes, central nervous system, kidneys, and liver. Mice with T cell-specific deletion of any two Zfp36 genes were protected from this spontaneous syndrome. Triply deficient T cells overproduced proinflammatory cytokines, including IFN-γ, TNF, and GM-CSF, due to increased mRNA stability of these transcripts. Unexpectedly, T cell-specific deletion of both Zfp36l1 and Zfp36l2 rendered mice resistant to experimental autoimmune encephalomyelitits due to failed priming of antigen-specific CD4+ T cells. ZFP36L1 and ZFP36L2 double-deficient CD4+ T cells had poor proliferation during in vitro T helper cell polarization. Thus, the ZFP36 family redundantly regulates T cell quiescence at homeostasis, but ZFP36L1 and ZFP36L2 are specifically required for antigen-specific T cell clonal expansion.
Figures






References
-
- Anderson P, Post-transcriptional control of cytokine production. Nat Immunol 9, 353–359 (2008). - PubMed
-
- Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G, Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity 10, 387–398 (1999). - PubMed
-
- Hodge DL, Berthet C, Coppola V, Kastenmüller W, Buschman MD, Schaughency PM, Shirota H, Scarzello AJ, Subleski JJ, Anver MR, Ortaldo JR, Lin F, Reynolds DA, Sanford ME, Kaldis P, Tessarollo L, Klinman DM, Young HA, IFN-gamma AU-rich element removal promotes chronic IFN-gamma expression and autoimmunity in mice. J Autoimmun 53, 33–45 (2014). - PMC - PubMed
-
- Matsushita K, Takeuchi O, Standley DM, Kumagai Y, Kawagoe T, Miyake T, Satoh T, Kato H, Tsujimura T, Nakamura H, Akira S, Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature 458, 1185–1190 (2009). - PubMed
-
- Uehata T, Iwasaki H, Vandenbon A, Matsushita K, Hernandez-Cuellar E, Kuniyoshi K, Satoh T, Mino T, Suzuki Y, Standley DM, Tsujimura T, Rakugi H, Isaka Y, Takeuchi O, Akira S, Malt1-induced cleavage of regnase-1 in CD4(+) helper T cells regulates immune activation. Cell 153, 1036–1049 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

