Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75
- PMID: 36270286
- PMCID: PMC9531665
- DOI: 10.1016/j.chom.2022.09.018
Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75
Abstract
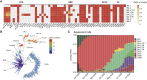
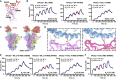
Recently emerged SARS-CoV-2 Omicron subvariant, BA.2.75, displayed a growth advantage over circulating BA.2.38, BA.2.76, and BA.5 in India. However, the underlying mechanisms for enhanced infectivity, especially compared with BA.5, remain unclear. Here, we show that BA.2.75 exhibits substantially higher affinity for host receptor angiotensin-converting enzyme 2 (ACE2) than BA.5 and other variants. Structural analyses of BA.2.75 spike shows its decreased thermostability and increased frequency of the receptor binding domain (RBD) in the "up" conformation under acidic conditions, suggesting enhanced low-pH-endosomal cell entry. Relative to BA.4/BA.5, BA.2.75 exhibits reduced evasion of humoral immunity from BA.1/BA.2 breakthrough-infection convalescent plasma but greater evasion of Delta breakthrough-infection convalescent plasma. BA.5 breakthrough-infection plasma also exhibits weaker neutralization against BA.2.75 than BA.5, mainly due to BA.2.75's distinct neutralizing antibody (NAb) escape pattern. Antibody therapeutics Evusheld and Bebtelovimab remain effective against BA.2.75. These results suggest BA.2.75 may prevail after BA.4/BA.5, and its increased receptor-binding capability could support further immune-evasive mutations.
Keywords: BA.2.75; COVID-19; Omicron; SARS-CoV-2; cryo-EM structure; immune evasion; neutralizing antibody; spike glycoprotein.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests X.S.X. and Y.C. are inventors on the provisional patent applications of BD series antibodies, which includes BD30-604 (DXP-604), BD55-5840 (SA58), and BD55-5514 (SA55). X.S.X. and Y.C. are founders of Singlomics Biopharmaceuticals.
Figures






References
-
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. Phenix: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
-
- Bruel T., Hadjadj J., Maes P., Planas D., Seve A., Staropoli I., Guivel-Benhassine F., Porrot F., Bolland W.H., Nguyen Y., et al. Serum neutralization of SARS-CoV-2 Omicron sublineages BA.1 and BA.2 in patients receiving monoclonal antibodies. Nat. Med. 2022;28:1297–1302. doi: 10.1038/s41591-022-01792-5. - DOI - PubMed
-
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

