Hypothesis-driven dragging of transcriptomic data to analyze proven targeted pathways in Rhinella arenarum larvae exposed to organophosphorus pesticides
- PMID: 36271284
- PMCID: PMC9587056
- DOI: 10.1038/s41598-022-21748-6
Hypothesis-driven dragging of transcriptomic data to analyze proven targeted pathways in Rhinella arenarum larvae exposed to organophosphorus pesticides
Abstract
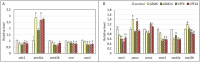
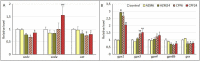
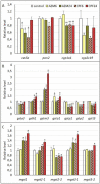
Transcriptional analysis of the network of transcription regulators and target pathways in exposed organisms may be a hard task when their genome remains unknown. The development of hundreds of qPCR assays, including primer design and normalization of the results with the appropriate housekeeping genes, seems an unreachable task. Alternatively, we took advantage of a whole transcriptome study on Rhinella arenarum larvae exposed to the organophosphorus pesticides azinphos-methyl and chlorpyrifos to evaluate the transcriptional effects on a priori selected groups of genes. This approach allowed us to evaluate the effects on hypothesis-selected pathways such as target esterases, detoxifying enzymes, polyamine metabolism and signaling, and regulatory pathways modulating them. We could then compare the responses at the transcriptional level with previously described effects at the enzymatic or metabolic levels to obtain global insight into toxicity-response mechanisms. The effects of both pesticides on the transcript levels of these pathways could be considered moderate, while chlorpyrifos-induced responses were more potent and earlier than those elicited by azinphos-methyl. Finally, we inferred a prevailing downregulation effect of pesticides on signaling pathways and transcription factor transcripts encoding products that modulate/control the polyamine and antioxidant response pathways. We also tested and selected potential housekeeping genes based on those reported for other species. These results allow us to conduct future confirmatory studies on pesticide modulation of gene expression in toad larvae.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
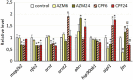


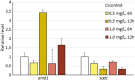

References
-
- Anguiano, O.L. & Pechen D’Angelo, A.M. Provincia de Río Negro y provincia de Neuquén in La problemática de los agroquímicos y sus envases, su incidencia en la salud de los trabajadores, la población expuesta y el ambiente. Estudio colaborativo multicéntrico (ed. Nonna, S., Secretaría de Ambiente y Desarrollo Sustentable) 181-210 ISBN 978-987-96256-7-5 (Ministerio de Salud República Argentina, 2007).
-
- Alvarez M, Du Mortier C, Jaureguiberry S, Venturino A. Joint probabilistic analysis of risk for aquatic species and exceedence frequency for the agricultural use of chlorpyrifos in the Pampean region Argentina. Environ. Toxicol. Chem. 2019;38:1748–1755. - PubMed
-
- Loewy RM, Kirs V, Carvajal LG, Venturino A, Pechen D’Angelo AM. Groundwater contamination by azinphos methyl in the Northern Patagonic region (Argentina) Sci. Total Environ. 1999;225:211–218. - PubMed
-
- Rosenbaum EA, et al. Response of biomarkers in amphibian larvae to in situ exposures in a fruit-producing region in North Patagonia Argentina. Environ. Toxicol. Chem. 2012;31:2311–2317. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

