Development and characterization of a new monoclonal antibody against SARS-CoV-2 NSP12 (RdRp)
- PMID: 36271490
- PMCID: PMC9874566
- DOI: 10.1002/jmv.28246
Development and characterization of a new monoclonal antibody against SARS-CoV-2 NSP12 (RdRp)
Abstract
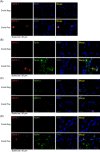
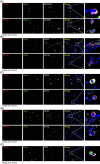
SARS-CoV-2 NSP12, the viral RNA-dependent RNA polymerase (RdRp), is required for viral replication and is a therapeutic target to treat COVID-19. To facilitate research on SARS-CoV-2 NSP12 protein, we developed a rat monoclonal antibody (CM12.1) against the NSP12 N-terminus that can facilitate functional studies. Immunoblotting and immunofluorescence assay (IFA) confirmed the specific detection of NSP12 protein by this antibody for cells overexpressing the protein. Although NSP12 is generated from the ORF1ab polyprotein, IFA of human autopsy COVID-19 lung samples revealed NSP12 expression in only a small fraction of lung cells including goblet, club-like, vascular endothelial cells, and a range of immune cells, despite wide-spread tissue expression of spike protein antigen. Similar studies using in vitro infection also generated scant protein detection in cells with established virus replication. These results suggest that NSP12 may have diminished steady-state expression or extensive posttranslation modifications that limit antibody reactivity during SARS-CoV-2 replication.
Keywords: COVID-19; NSP12; RNA-dependent RNA polymerase, RdRp; SARS-CoV-2; monoclonal antibody.
© 2022 Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

