Fungal names: a comprehensive nomenclatural repository and knowledge base for fungal taxonomy
- PMID: 36271801
- PMCID: PMC9825588
- DOI: 10.1093/nar/gkac926
Fungal names: a comprehensive nomenclatural repository and knowledge base for fungal taxonomy
Abstract
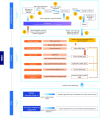
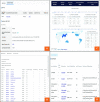
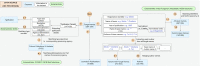
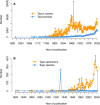
Fungal taxonomy is a complex and rapidly changing subject, which makes proper naming of fungi challenging for taxonomists. A registration platform with a standardized and information-integrated database is a powerful tool for efficient research on fungal taxonomy. Fungal Names (FN, https://nmdc.cn/fungalnames/; launched in 2011) is one of the three official fungal nomenclatural repositories authorized by the International Nomenclature Committee for Fungi (NCF). Currently, FN includes >567 000 taxon names from >10 000 related journals and books published since 1596 and covers >147 000 collection records of type specimens/illustrations from >5000 preserving agencies. FN is also a knowledge base that integrates nomenclature information with specimens, culture collections and herbaria/fungaria, publications and taxonomists, and represents a summary of the history and recent advances in fungal taxonomy. Published fungal names are categorized based on well-accepted nomenclature rules and can be readily searched with different keywords and strategies. In combination with a standardized name checking tool and a sequence alignment-based identification package, FN makes the registration and typification of nomenclatural novelties of fungi convenient and accurate.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Turland N., Wiersema J., Barrie F., Greuter W., Hawksworth D., Herendeen P., Knapp S., Kusber W.-H., Li D.-Z., Marhold K.et al. .. International Code of Nomenclature for Algae, Fungi, and Plants (Shenzhen Code) adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017. 2018; Glashütten: Koeltz Botanical Books.
-
- McNeill J., Turland N.J.. Major changes to the code of nomenclature—melbourne. Taxon. 2011; 60:1495–1497.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

