EV-ADD, a database for EV-associated DNA in human liquid biopsy samples
- PMID: 36271888
- PMCID: PMC9587709
- DOI: 10.1002/jev2.12270
EV-ADD, a database for EV-associated DNA in human liquid biopsy samples
Abstract
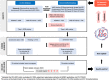
Extracellular vesicles (EVs) play a key role in cellular communication both in physiological conditions and in pathologies such as cancer. Emerging evidence has shown that EVs are active carriers of molecular cargo (e.g. protein and nucleic acids) and a powerful source of biomarkers and targets. While recent studies on EV-associated DNA (EV-DNA) in human biofluids have generated a large amount of data, there is currently no database that catalogues information on EV-DNA. To fill this gap, we have manually curated a database of EV-DNA data derived from human biofluids (liquid biopsy) and in-vitro studies, called the Extracellular Vesicle-Associated DNA Database (EV-ADD). This database contains validated experimental details and data extracted from peer-reviewed published literature. It can be easily queried to search for EV isolation methods and characterization, EV-DNA isolation techniques, quality validation, DNA fragment size, volume of starting material, gene names and disease context. Currently, our database contains samples representing 23 diseases, with 13 different types of EV isolation techniques applied on eight different human biofluids (e.g. blood, saliva). In addition, EV-ADD encompasses EV-DNA data both representing the whole genome and specifically including oncogenes, such as KRAS, EGFR, BRAF, MYC, and mitochondrial DNA (mtDNA). An EV-ADD data metric system was also integrated to assign a compliancy score to the MISEV guidelines based on experimental parameters reported in each study. While currently available databases document the presence of proteins, lipids, RNA and metabolites in EVs (e.g. Vesiclepedia, ExoCarta, ExoBCD, EVpedia, and EV-TRACK), to the best of our knowledge, EV-ADD is the first of its kind to compile all available EV-DNA datasets derived from human biofluid samples. We believe that this database provides an important reference resource on EV-DNA-based liquid biopsy research, serving as a learning tool and to showcase the latest developments in the EV-DNA field. EV-ADD will be updated yearly as newly published EV-DNA data becomes available and it is freely available at www.evdnadatabase.com.
Keywords: EV-ADD; EV-DNA; cfDNA; database; extracellular vesicles; liquid biopsy.
© 2022 The Authors. Journal of Extracellular Vesicles published by Wiley Periodicals, LLC on behalf of the International Society for Extracellular Vesicles.
Figures

References
-
- Abramowicz, A. , Widlak, P. , & Pietrowska, M. (2016). Proteomic analysis of exosomal cargo: The challenge of high purity vesicle isolation. Molecular Biosystems, 12, 1407–1419. - PubMed
-
- Allenson, K. , Castillo, J. , San Lucas, F. A. , Scelo, G. , Kim, D. U. , Bernard, V. , Davis, G. , Kumar, T. , Katz, M. , Overman, M. J. , Foretova, L. , Fabianova, E. , Holcatova, I. , Janout, V. , Meric‐Bernstam, F. , Gascoyne, P. , Wistuba, I. , Varadhachary, G. , Brennan, P. , … Alvarez, H. (2017). High prevalence of mutant KRAS in circulating exosome‐derived DNA from early‐stage pancreatic cancer patients. Annals of Oncology, 28, 741–747. - PMC - PubMed
-
- Arraud, N. , Linares, R. , Tan, S. , Gounou, C. , Pasquet, J. M. , Mornet, S. , & Brisson, A. R. (2014). Extracellular vesicles from blood plasma: Determination of their morphology, size, phenotype and concentration. Journal of Thrombosis and Haemostasis, 12, 614–627. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

