Adaptations to high pressure of Nautilia sp. strain PV-1, a piezophilic Campylobacterium (aka Epsilonproteobacterium) isolated from a deep-sea hydrothermal vent
- PMID: 36271901
- PMCID: PMC10092268
- DOI: 10.1111/1462-2920.16256
Adaptations to high pressure of Nautilia sp. strain PV-1, a piezophilic Campylobacterium (aka Epsilonproteobacterium) isolated from a deep-sea hydrothermal vent
Abstract
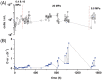
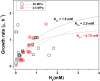
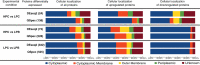
Physiological and gene expression studies of deep-sea bacteria under pressure conditions similar to those experienced in their natural habitat are critical for understanding growth kinetics and metabolic adaptations to in situ conditions. The Campylobacterium (aka Epsilonproteobacterium) Nautilia sp. strain PV-1 was isolated from hydrothermal fluids released from an active deep-sea hydrothermal vent at 9° N on the East Pacific Rise. Strain PV-1 is a piezophilic, moderately thermophilic, chemolithoautotrophic anaerobe that conserves energy by coupling the oxidation of hydrogen to the reduction of nitrate or elemental sulfur. Using a high-pressure-high temperature continuous culture system, we established that strain PV-1 has the shortest generation time of all known piezophilic bacteria and we investigated its protein expression pattern in response to different hydrostatic pressure regimes. Proteogenomic analyses of strain PV-1 grown at 20 and 5 MPa showed that pressure adaptation is not restricted to stress response or homeoviscous adaptation but extends to enzymes involved in central metabolic pathways. Protein synthesis, motility, transport, and energy metabolism are all affected by pressure, although to different extents. In strain PV-1, low-pressure conditions induce the synthesis of phage-related proteins and an overexpression of enzymes involved in carbon fixation.
© 2022 The Authors. Environmental Microbiology published by Applied Microbiology International and John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures




References
-
- Alain, K. , Callac, N. , Guégan, M. , Lesongeur, F. , Crassous, P. , Cambon‐Bonavita, M.‐A. et al. (2009) Nautilia abyssi sp. nov., a thermophilic, chemolithoautotrophic, sulfur‐reducing bacterium isolated from an East Pacific Rise hydrothermal vent. International Journal of Systematic and Evolutionary Microbiology, 59, 1310–1315. Available from: 10.1099/ijs.0.005454-0 - DOI - PubMed
-
- Alain, K. , Marteinsson, V.T. , Miroshnichenko, M.L. , Bonch‐Osmolovskaya, E.A. , Prieur, D. & Birrien, J.‐L. (2002) Marinitoga piezophila sp. nov., a rod‐shaped, thermo‐piezophilic bacterium isolated under high hydrostatic pressure from a deep‐sea hydrothermal vent. International Journal of Systematic and Evolutionary Microbiology, 52, 1331–1339. Available from: 10.1099/00207713-52-4-1331 - DOI - PubMed
-
- Amrani, A. , Bergon, A. , Holota, H. , Tamburini, C. , Garel, M. , Ollivier, B. et al. (2014) Transcriptomics reveal several gene expression patterns in the Piezophile Desulfovibrio hydrothermalis in response to hydrostatic pressure. PLoS One, 9, e106831. Available from: 10.1371/journal.pone.0106831 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

