Expanded analysis of high-grade astrocytoma with piloid features identifies an epigenetically and clinically distinct subtype associated with neurofibromatosis type 1
- PMID: 36271929
- PMCID: PMC9844520
- DOI: 10.1007/s00401-022-02513-5
Expanded analysis of high-grade astrocytoma with piloid features identifies an epigenetically and clinically distinct subtype associated with neurofibromatosis type 1
Abstract
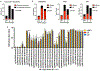
High-grade astrocytoma with piloid features (HGAP) is a recently recognized glioma type whose classification is dependent on its global epigenetic signature. HGAP is characterized by alterations in the mitogen-activated protein kinase (MAPK) pathway, often co-occurring with CDKN2A/B homozygous deletion and/or ATRX mutation. Experience with HGAP is limited and to better understand this tumor type, we evaluated an expanded cohort of patients (n = 144) with these tumors, as defined by DNA methylation array testing, with a subset additionally evaluated by next-generation sequencing (NGS). Among evaluable cases, we confirmed the high prevalence CDKN2A/B homozygous deletion, and/or ATRX mutations/loss in this tumor type, along with a subset showing NF1 alterations. Five of 93 (5.4%) cases sequenced harbored TP53 mutations and RNA fusion analysis identified a single tumor containing an NTRK2 gene fusion, neither of which have been previously reported in HGAP. Clustering analysis revealed the presence of three distinct HGAP subtypes (or groups = g) based on whole-genome DNA methylation patterns, which we provisionally designated as gNF1 (n = 18), g1 (n = 72), and g2 (n = 54) (median ages 43.5 years, 47 years, and 32 years, respectively). Subtype gNF1 is notable for enrichment with patients with Neurofibromatosis Type 1 (33.3%, p = 0.0008), confinement to the posterior fossa, hypermethylation in the NF1 enhancer region, a trend towards decreased progression-free survival (p = 0.0579), RNA processing pathway dysregulation, and elevated non-neoplastic glia and neuron cell content (p < 0.0001 and p < 0.0001, respectively). Overall, our expanded cohort broadens the genetic, epigenetic, and clinical phenotype of HGAP and provides evidence for distinct epigenetic subtypes in this tumor type.
Keywords: DNA Methylation; HGAP; High-grade astrocytoma with piloid features; NF1; Neurofibromatosis type 1.
© 2022. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Figures




References
-
- Alexander BM, Trippa L, Gaffey S, Arrillaga-Romany IC, Lee EQ, Rinne ML et al. (2019) Individualized Screening Trial of Innovative Glioblastoma Therapy (INSIGhT): a Bayesian adaptive platform trial to develop precision medicines for patients with glioblastoma. JCO Precis Oncol. 10.1200/PO.18.00071 - DOI - PMC - PubMed
-
- Bates D, Machler M, Bolker B, Walker S (2015) Fitting linear mixe-effects models using lme4. J Stat Softw 67:1–48. 10.18637/jss.v067.i01 - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

