Hybridization Chain Reaction Fluorescence In Situ Hybridization (HCR-FISH) in Ambystoma mexicanum Tissue
- PMID: 36272070
- PMCID: PMC10949069
- DOI: 10.1007/978-1-0716-2659-7_6
Hybridization Chain Reaction Fluorescence In Situ Hybridization (HCR-FISH) in Ambystoma mexicanum Tissue
Abstract
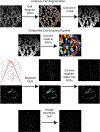
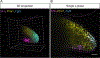
In situ hybridization is a standard procedure for visualizing mRNA transcripts in tissues. The recent adoption of fluorescent probes and new signal amplification methods have facilitated multiplexed RNA imaging in tissue sections and whole tissues. Here we present protocols for multiplexed hybridization chain reaction fluorescence in situ hybridization (HCR-FISH) staining, imaging, cell segmentation, and mRNA quantification in regenerating axolotl tissue sections. We also present a protocol for whole-mount staining and imaging of developing axolotl limbs.
Keywords: Axolotl; Hybridization chain reaction; In situ hybridization; Limb development; Limb regeneration.
© 2023. The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature.
Conflict of interest statement
Figures
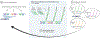


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

