From thymus to tissues and tumors: A review of T-cell biology
- PMID: 36272581
- PMCID: PMC9825672
- DOI: 10.1016/j.jaci.2022.10.011
From thymus to tissues and tumors: A review of T-cell biology
Abstract
T cells are critical orchestrators of the adaptive immune response that optimally eliminate a specific pathogen. Aberrant T-cell development and function are implicated in a broad range of human disease including immunodeficiencies, autoimmune diseases, and allergic diseases. Accordingly, therapies targeting T cells and their effector cytokines have markedly improved the care of patients with immune dysregulatory diseases. Newer discoveries concerning T-cell-mediated antitumor immunity and T-cell exhaustion have further prompted development of highly effective and novel treatment modalities for malignancies, including checkpoint inhibitors and antigen-reactive T cells. Recent discoveries are also uncovering the depth and variability of T-cell phenotypes: while T cells have long been described using a subset-based classification system, next-generation sequencing technologies suggest an astounding degree of complexity and heterogeneity at the single-cell level.
Keywords: CD4; CD8; T cells; adaptive immunity.
Published by Elsevier Inc.
Conflict of interest statement
Figures
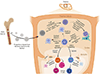


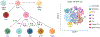
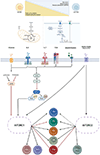

References
-
- June CH, O'Connor RS, Kawalekar OU, Ghassemi S, Milone MC. CAR T cell immunotherapy for human cancer. Science 2018; 359:1361–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

