Sex Differences in Genomic Features of Hepatitis B-Associated Hepatocellular Carcinoma With Distinct Antitumor Immunity
- PMID: 36272708
- PMCID: PMC9772570
- DOI: 10.1016/j.jcmgh.2022.10.009
Sex Differences in Genomic Features of Hepatitis B-Associated Hepatocellular Carcinoma With Distinct Antitumor Immunity
Abstract
Background & aims: Aflatoxin exposure increases the risk for hepatocellular carcinoma (HCC) in hepatitis B virus (HBV)-infected individuals, particularly males. We investigated sex-based differences in the HCC genome and antitumor immunity.
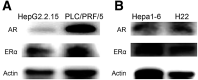
Methods: Whole-genome, whole-exome, and RNA sequencing were performed on 101 HCC patient samples (47 males, 54 females) that resulted from HBV infection and aflatoxin exposure from Qidong. Androgen on the expression of aflatoxin metabolism-related genes and nonhomologous DNA end joining (NHEJ) factors were examined in HBV-positive HCC cell lines, and further tested in tumor-bearing syngeneic mice.
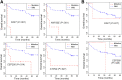
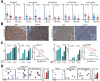
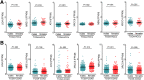
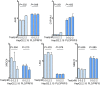
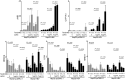
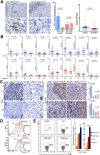
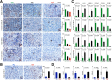
Results: Qidong HCC differed between males and females in genomic landscape and transcriptional dysfunction pathways. Compared with females, males expressed higher levels of aflatoxin metabolism-related genes, such as AHR and CYP1A1, and lower levels of NHEJ factors, such as XRCC4, LIG4, and MRE11, showed a signature of up-regulated type I interferon signaling/response and repressed antitumor immunity. Treatment with AFB1 in HBV-positive cells, the addition of 2 nmol/L testosterone to cultures significantly increased the expression of aflatoxin metabolism-related genes, but reduced NHEJ factors, resulting in more nuclear DNA leakage into cytosol to activate cGAS-STING. In syngeneic tumor-bearing mice that were administrated tamoxifen daily via oral gavage, favorable androgen signaling repressed NHEJ factor expression and activated cGAS-STING in tumors, increasing T-cell infiltration and improving anti-programmed cell death protein 1 treatment effect.
Conclusions: Androgen signaling in the context of genotoxic stress repressed DNA damage repair. The alteration caused more nuclear DNA leakage into cytosol to activate the cGAS-STING pathway, which increased T-cell infiltration into tumor mass and improved anti-programmed cell death protein 1 immunotherapy in HCCs.
Keywords: Aflatoxin; DNA Double-Strand Break; HBV; Immune Checkpoints; Sex Hormones.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Figures









References
-
- Wang S.H., Yeh S.H., Lin W.H., Yeh K.H., Yuan Q., Xia N.S., Chen D.S., Chen P.J. Estrogen receptor alpha represses transcription of HBV genes via interaction with hepatocyte nuclear factor 4alpha. Gastroenterology. 2012;142:989–998 e4. - PubMed
-
- Nault J.C., Martin Y., Caruso S., Hirsch T.Z., Bayard Q., Calderaro J., Charpy C., Copie-Bergman C., Ziol M., Bioulac-Sage P., Couchy G., Blanc J.F., Nahon P., Amaddeo G., Ganne-Carrie N., Morcrette G., Chiche L., Duvoux C., Faivre S., Laurent A., Imbeaud S., Rebouissou S., Llovet J.M., Seror O., Letouze E., Zucman-Rossi J. Clinical impact of genomic diversity from early to advanced hepatocellular carcinoma. Hepatology. 2020;71:164–182. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

