Feasibility of the optimal cerebral perfusion pressure value identification without a delay that is too long
- PMID: 36272984
- PMCID: PMC9588030
- DOI: 10.1038/s41598-022-22566-6
Feasibility of the optimal cerebral perfusion pressure value identification without a delay that is too long
Abstract
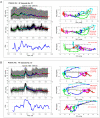
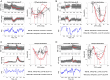
Optimal cerebral perfusion pressure (CPPopt)-targeted treatment of traumatic brain injury (TBI) patients requires 2-8 h multi-modal monitoring data accumulation to identify CPPopt value for individual patient. Minimizing the time required for monitoring data accumulation is needed to improve the efficacy of CPPopt-targeted therapy. A retrospective analysis of multimodal physiological monitoring data from 87 severe TBI patients was performed by separately representing cerebrovascular autoregulation (CA) indices in relation to CPP, arterial blood pressure (ABP), and intracranial pressure (ICP) to improve the existing CPPopt identification algorithms. Machine learning (ML)-based algorithms were developed for automatic identification of informative data segments that were used for reliable CPPopt, ABPopt, ICPopt and the lower/upper limits of CA (LLCA/ULCA) identification. The reference datasets of the informative data segments and, artifact-distorted segments, and the datasets of different clinical situations were used for training the ML-based algorithms, allowing us to choose the appropriate individualized CPP-, ABP- or ICP-guided management for 79% of the full monitoring time for the studied population. The developed ML-based algorithms allow us to recognize informative physiological ABP/ICP variations within 24 min intervals with an accuracy up to 79% (compared to the initial accuracy of 74%) and use these segments for timely optimal value identification or CA limits determination in CPP, ABP or ICP data. Prospective clinical studies are needed to prove the efficiency of the developed algorithms.
© 2022. The Author(s).
Conflict of interest statement
V.Pe., M.D., E.C., S.R. and K.Be. received a research grant from the Research Council of Lithuania (Grant No. MIP-20-216). V.Pe., M.K. and E.C. received a research grant from the Research and Innovation Fund of Kaunas University of Technology (Grant No. PP54/203). A.T. and T.T. received a research grant from the Research Fund of the Lithuanian University of Health Sciences (Grant No. PP54/203). S.K. received a research grant from the EU Structural Funds, Promotion of Post-Doctoral Fellowships (Grant No. 09.3.3-LMT-K-712-19-0023). V.Pe., M.D., E.C., M.K., A.R., A.P. and T.T. are coauthors of the developed ML-based software tool for ICU multimodal monitoring data analysis and CPPopt, ABPopt, or LLCA/ULCA identification. A.R., S.V., A.P., V.Pu. and K.Bo. declare no potential conflict of interest.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

