Patient-specific modeling of stroma-mediated chemoresistance of pancreatic cancer using a three-dimensional organoid-fibroblast co-culture system
- PMID: 36273171
- PMCID: PMC9588250
- DOI: 10.1186/s13046-022-02519-7
Patient-specific modeling of stroma-mediated chemoresistance of pancreatic cancer using a three-dimensional organoid-fibroblast co-culture system
Abstract
Background: Cancer-associated fibroblasts (CAFs) are considered to play a fundamental role in pancreatic ductal adenocarcinoma (PDAC) progression and chemoresistance. Patient-derived organoids have demonstrated great potential as tumor avatars for drug response prediction in PDAC, yet they disregard the influence of stromal components on chemosensitivity.
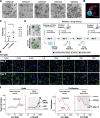
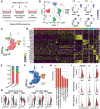
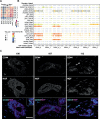
Methods: We established direct three-dimensional (3D) co-cultures of primary PDAC organoids and patient-matched CAFs to investigate the effect of the fibroblastic compartment on sensitivity to gemcitabine, 5-fluorouracil and paclitaxel treatments using an image-based drug assay. Single-cell RNA sequencing was performed for three organoid/CAF pairs in mono- and co-culture to uncover transcriptional changes induced by tumor-stroma interaction.
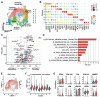
Results: Upon co-culture with CAFs, we observed increased proliferation and reduced chemotherapy-induced cell death of PDAC organoids. Single-cell RNA sequencing data evidenced induction of a pro-inflammatory phenotype in CAFs in co-cultures. Organoids showed increased expression of genes associated with epithelial-to-mesenchymal transition (EMT) in co-cultures and several potential receptor-ligand interactions related to EMT were identified, supporting a key role of CAF-driven induction of EMT in PDAC chemoresistance.
Conclusions: Our results demonstrate the potential of personalized PDAC co-cultures models not only for drug response profiling but also for unraveling the molecular mechanisms involved in the chemoresistance-supporting role of the tumor stroma.
Keywords: Cancer-associated fibroblasts; Drug screening; Pancreatic cancer; Patient-derived organoids; Personalized oncology.
© 2022. The Author(s).
Conflict of interest statement
J. Jabs was employed by DKFZ and Steinbeis GmbH during experimental work and analysis and at Merck Healthcare KGaA during writing of the manuscript. All other authors declare no conflict of interest.
Figures

References
-
- Ferlay J, Ervik M, Lam F, Colombet M, Mery L, Piñeros M, et al. Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer. 2020 [cited 2022 Mar 7]. Available from: https://gco.iarc.fr/today
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

