Statistical methods for cis-Mendelian randomization with two-sample summary-level data
- PMID: 36273411
- PMCID: PMC7614127
- DOI: 10.1002/gepi.22506
Statistical methods for cis-Mendelian randomization with two-sample summary-level data
Abstract
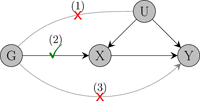
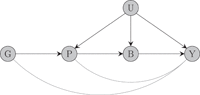
Mendelian randomization (MR) is the use of genetic variants to assess the existence of a causal relationship between a risk factor and an outcome of interest. Here, we focus on two-sample summary-data MR analyses with many correlated variants from a single gene region, particularly on cis-MR studies which use protein expression as a risk factor. Such studies must rely on a small, curated set of variants from the studied region; using all variants in the region requires inverting an ill-conditioned genetic correlation matrix and results in numerically unstable causal effect estimates. We review methods for variable selection and estimation in cis-MR with summary-level data, ranging from stepwise pruning and conditional analysis to principal components analysis, factor analysis, and Bayesian variable selection. In a simulation study, we show that the various methods have comparable performance in analyses with large sample sizes and strong genetic instruments. However, when weak instrument bias is suspected, factor analysis and Bayesian variable selection produce more reliable inferences than simple pruning approaches, which are often used in practice. We conclude by examining two case studies, assessing the effects of low-density lipoprotein-cholesterol and serum testosterone on coronary heart disease risk using variants in the HMGCR and SHBG gene regions, respectively.
Keywords: Bayesian variable selection; Mendelian randomization; correlated instruments; factor analysis; principal components analysis; pruning.
© 2022 The Authors. Genetic Epidemiology published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Allara, E. , Morani, G. , Carter, P. , Gkatzionis, A. , Zuber, V. , Foley, C. N. , Rees, J. M. , Mason, A. M. , Bell, S. , Gill, D. , Lindstrom, S. , Butterworth, A. S. , DiAngelantonio, E. , Peters, J. , Burgess, S. , & INVENT consortium (2019). Genetic determinants of lipids and cardiovascular disease outcomes: A wide‐angled Mendelian randomization investigation. Circulation. Genomic and precision medicine, 12(12), e002711. - PMC - PubMed
-
- Anderson, T. W. , & Rubin, H. (1949). Estimation of the parameters of a single equation in a complete system of stochastic equations. Annals of Mathematical Statistics, 20(1), 46–63.
-
- Angrist, J. D. , & Pischke, J.‐S. (2008). Mostly harmless econometrics: An empiricist's companion. Princeton University Press.
-
- Asimit, J. L. , Rainbow, D. B. , Fortune, M. D. , Grinberg, N. F. , Wicker, L. S. , & Wallace, C. (2019). Stochastic search and joint fine‐mapping increases accuracy and identifies previously unreported associations in immune‐mediated diseases. Nature Communications, 10. 10.1038/s41467-019-11271-0 - DOI - PMC - PubMed
-
- Bai, J. , & Ng, S. (2002). Determining the number of factors in approximate factor models. Econometrica, 70(1), 191–221.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

