High-resolution transcriptomics informs glial pathology in human temporal lobe epilepsy
- PMID: 36274170
- PMCID: PMC9590125
- DOI: 10.1186/s40478-022-01453-1
High-resolution transcriptomics informs glial pathology in human temporal lobe epilepsy
Erratum in
-
Correction: High-resolution transcriptomics informs glial pathology in human temporal lobe epilepsy.Acta Neuropathol Commun. 2022 Nov 28;10(1):171. doi: 10.1186/s40478-022-01479-5. Acta Neuropathol Commun. 2022. PMID: 36443832 Free PMC article. No abstract available.
Abstract
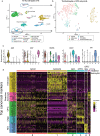
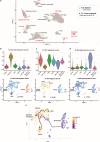
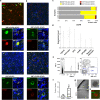
The pathophysiology of epilepsy underlies a complex network dysfunction between neurons and glia, the molecular cell type-specific contributions of which remain poorly defined in the human disease. In this study, we validated a method that simultaneously isolates neuronal (NEUN +), astrocyte (PAX6 + NEUN-), and oligodendroglial progenitor (OPC) (OLIG2 + NEUN-) enriched nuclei populations from non-diseased, fresh-frozen human neocortex and then applied it to characterize the distinct transcriptomes of such populations isolated from electrode-mapped temporal lobe epilepsy (TLE) surgical samples. Nuclear RNA-seq confirmed cell type specificity and informed both common and distinct pathways associated with TLE in astrocytes, OPCs, and neurons. Compared to postmortem control, the transcriptome of epilepsy astrocytes showed downregulation of mature astrocyte functions and upregulation of development-related genes. To gain further insight into glial heterogeneity in TLE, we performed single cell transcriptomics (scRNA-seq) on four additional human TLE samples. Analysis of the integrated TLE dataset uncovered a prominent subpopulation of glia that express a hybrid signature of both reactive astrocyte and OPC markers, including many cells with a mixed GFAP + OLIG2 + phenotype. A further integrated analysis of this TLE scRNA-seq dataset and a previously published normal human temporal lobe scRNA-seq dataset confirmed the unique presence of hybrid glia only in TLE. Pseudotime analysis revealed cell transition trajectories stemming from this hybrid population towards both OPCs and reactive astrocytes. Immunofluorescence studies in human TLE samples confirmed the rare presence of GFAP + OLIG2 + glia, including some cells with proliferative activity, and functional analysis of cells isolated directly from these samples disclosed abnormal neurosphere formation in vitro. Overall, cell type-specific isolation of glia from surgical epilepsy samples combined with transcriptomic analyses uncovered abnormal glial subpopulations with de-differentiated phenotype, motivating further studies into the dysfunctional role of reactive glia in temporal lobe epilepsy.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this article.
Figures



References
-
- Bell GS, Sander JW: The epidemiology of epilepsy: the size of the problem. Seizure 2001, 10(4):306–314; quiz 315–306. - PubMed
-
- Bien CG, Kurthen M, Baron K, Lux S, Helmstaedter C, Schramm J, Elger CE. Long-term seizure outcome and antiepileptic drug treatment in surgically treated temporal lobe epilepsy patients: a controlled study. Epilepsia. 2001;42(11):1416–1421. - PubMed
-
- Legido A, Katsetos CD. Experimental studies in epilepsy: immunologic and inflammatory mechanisms. Seminars in pediatric neurology. 2014;21(3):197–206. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

