Metatranscriptomic and metataxonomic insights into the ultra-small microbiome of the Korean fermented vegetable, kimchi
- PMID: 36274711
- PMCID: PMC9581167
- DOI: 10.3389/fmicb.2022.1026513
Metatranscriptomic and metataxonomic insights into the ultra-small microbiome of the Korean fermented vegetable, kimchi
Abstract
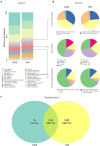
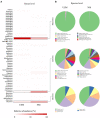
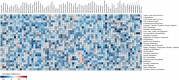
Presently, pertinent information on the ultra-small microbiome (USM) in fermented vegetables is still lacking. This study analyzed the metatranscriptome and metataxonome for the USM of kimchi. Tangential flow filtration was used to obtain a USM with a size of 0.2 μm or less from kimchi. The microbial diversity in the USM was compared with that of the normal microbiome (NM). Alpha diversity was higher in the USM than in NM, and the diversity of bacterial members of the NM was higher than that of the USM. At the phylum level, both USM and NM were dominated by Firmicutes. At the genus level, the USM and NM were dominated by Lactobacillus, Leuconostoc, and Weissella, belonging to lactic acid bacteria. However, as alpha diversity is higher in the USM than in the NM, the genus Akkermansia, belonging to the phylum Verrucomicrobia, was detected only in the USM. Compared to the NM, the USM showed a relatively higher ratio of transcripts related to "protein metabolism," and the USM was suspected to be involved with the viable-but-nonculturable (VBNC) state. When comparing the sub-transcripts related to the "cell wall and capsule" of USM and NM, USM showed a proportion of transcripts suspected of being VBNC. In addition, the RNA virome was also identified, and both the USM and NM were confirmed to be dominated by pepper mild mottle virus (PMMoV). Additionally, the correlation between metataxonome and metatranscriptome identified USM and NM was estimated, however, only limited correlations between metataxonome and metatranscriptome were estimated. This study provided insights into the relationship between the potential metabolic activities of the USM of kimchi and the NM.
Keywords: kimchi; metataxonome; metatranscriptome; tangential flow filtration; ultra-small microbiome.
Copyright © 2022 Lee, Yoon, Dang, Yun, Jeong, Kim, Bae and Ha.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Presence of an ultra-small microbiome in fermented cabbages.PeerJ. 2023 Jul 17;11:e15680. doi: 10.7717/peerj.15680. eCollection 2023. PeerJ. 2023. PMID: 37483986 Free PMC article.
-
The Microbial Diversity of Non-Korean Kimchi as Revealed by Viable Counting and Metataxonomic Sequencing.Foods. 2020 Oct 29;9(11):1568. doi: 10.3390/foods9111568. Foods. 2020. PMID: 33137924 Free PMC article.
-
Unraveling microbial fermentation features in kimchi: from classical to meta-omics approaches.Appl Microbiol Biotechnol. 2020 Sep;104(18):7731-7744. doi: 10.1007/s00253-020-10804-8. Epub 2020 Aug 4. Appl Microbiol Biotechnol. 2020. PMID: 32749526 Review.
-
Bacterial diversity of baechu-kimchi with seafood based on culture-independent investigations.Food Sci Biotechnol. 2023 Nov 29;33(7):1661-1670. doi: 10.1007/s10068-023-01471-2. eCollection 2024 Jun. Food Sci Biotechnol. 2023. PMID: 38623433 Free PMC article.
-
Kimchi microflora: history, current status, and perspectives for industrial kimchi production.Appl Microbiol Biotechnol. 2014 Mar;98(6):2385-93. doi: 10.1007/s00253-014-5513-1. Epub 2014 Jan 14. Appl Microbiol Biotechnol. 2014. PMID: 24419800 Review.
Cited by
-
The Weissella and Periweissella genera: up-to-date taxonomy, ecology, safety, biotechnological, and probiotic potential.Front Microbiol. 2023 Dec 11;14:1289937. doi: 10.3389/fmicb.2023.1289937. eCollection 2023. Front Microbiol. 2023. PMID: 38169702 Free PMC article. Review.
-
Presence of an ultra-small microbiome in fermented cabbages.PeerJ. 2023 Jul 17;11:e15680. doi: 10.7717/peerj.15680. eCollection 2023. PeerJ. 2023. PMID: 37483986 Free PMC article.
-
Lactic acid bacteria in Asian fermented foods and their beneficial roles in human health.Food Sci Biotechnol. 2024 Jun 17;33(9):2021-2033. doi: 10.1007/s10068-024-01634-9. eCollection 2024 Jul. Food Sci Biotechnol. 2024. PMID: 39130665 Free PMC article. Review.
-
Metatranscriptomics for Understanding the Microbiome in Food and Nutrition Science.Metabolites. 2025 Mar 10;15(3):185. doi: 10.3390/metabo15030185. Metabolites. 2025. PMID: 40137150 Free PMC article. Review.
-
The Rising Role of Omics and Meta-Omics in Table Olive Research.Foods. 2023 Oct 15;12(20):3783. doi: 10.3390/foods12203783. Foods. 2023. PMID: 37893676 Free PMC article. Review.
References
-
- Bikel S., Valdez-Lara A., Cornejo-Granados F., Rico K., Canizales-Quinteros S., Soberón X., et al. . (2015). Combining metagenomics, metatranscriptomics and viromics to explore novel microbial interactions: towards a systems-level understanding of human microbiome. Comput. Struct. Biotechnol. J. 13, 390–401. doi: 10.1016/j.csbj.2015.06.001, PMID: - DOI - PMC - PubMed
-
- Bousquet J., Anto J. M., Czarlewski W., Haahtela T., Fonseca S. C., Iaccarino G., et al. . (2021). Cabbage and fermented vegetables: from death rate heterogeneity in countries to candidates for mitigation strategies of severe COVID-19. Allergy 76, 735–750. doi: 10.1111/all.14549, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

