Molecular dynamics study of tropical calcific pancreatitis (TCP) associated calcium-sensing receptor single nucleotide variation
- PMID: 36275616
- PMCID: PMC9581290
- DOI: 10.3389/fmolb.2022.982831
Molecular dynamics study of tropical calcific pancreatitis (TCP) associated calcium-sensing receptor single nucleotide variation
Abstract
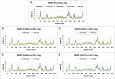
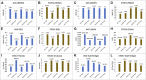
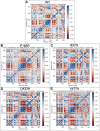
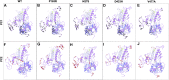
Tropical Calcific Pancreatitis (TCP) is a chronic non-alcoholic pancreatitis characterised by extensive calcification. The disease usually appears at a younger age and is more common in tropical regions. This disease's progression can lead to pancreatic diabetes, which can subsequently lead to pancreatic cancer. The CASR gene encodes a calcium-sensing receptor (CaSR), which is a GPCR protein of class C. It is expressed in the islets of Langerhans, the parathyroid gland, and other tissues. It primarily detects small gradients in circulating calcium concentrations and couples this information to intracellular signalling, which helps to regulate PTH (parathyroid hormone) secretion and mineral ion homeostasis. From co-leading insulin release, CaSR modulates ductal HCO3- secretion, Ca2+ concentration, cell-cell communication, β-cell proliferation, and intracellular Ca2+ release. In pancreatic cancer, the CaSR limits cell proliferation. TCP-related four novel missense mutations P163R, I427S, D433H and V477A, found in CaSR extracellular domain (ECD) protein, which were reported in the mutTCPdb Database (https://lms.snu.edu.in/mutTCPDB/index.php). P163R mutation occurs in ligand-binding domain 1 (LBD-1) of the CaSR ECD. To investigate the influence of these variations on protein function and structural activity multiple in-silico prediction techniques such as SIFT, PolyPhen, CADD scores, and other methods have been utilized. A 500 ns molecular dynamic simulation was performed on the CaSR ECD crystal structure and the corresponding mutated models. Furthermore, Principal Component Analysis (PCA) and Essential Dynamics analysis were used to forecast collective motions, thermodynamic stabilities, and the critical subspace crucial to CaSR functions. The results of molecular dynamic simulations showed that the mutations P163R, I427S, D433H, and V477A caused conformational changes and decreased the stability of protein structures. This study also demonstrates the significance of TCP associated mutations. As a result of our findings, we hypothesised that the investigated mutations may have an effect on the protein's structure and ability to interact with other molecules, which may be related to the protein's functional impairment.
Keywords: CaSR; calcium sensing receptor; molecular dynamics simulation; mutTCPdb; pancreatitis; single nucleotide variants; tropical calcific pancreatitis.
Copyright © 2022 Shrivastava, Mathur, Verma, Jayadev Magani, Vyas and Singh.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






Similar articles
-
mutTCPdb: a comprehensive database for genomic variants of a tropical country neglected disease-tropical calcific pancreatitis.Database (Oxford). 2018 Jan 1;2018:bay043. doi: 10.1093/database/bay043. Database (Oxford). 2018. PMID: 30053238 Free PMC article.
-
Novel mutations in the calcium sensing receptor gene in tropical chronic pancreatitis in India.Scand J Gastroenterol. 2008 Jan;43(1):117-21. doi: 10.1080/00365520701580413. Scand J Gastroenterol. 2008. PMID: 18938753
-
Structural, functional and molecular dynamics analysis of cathepsin B gene SNPs associated with tropical calcific pancreatitis, a rare disease of tropics.PeerJ. 2019 Oct 3;7:e7425. doi: 10.7717/peerj.7425. eCollection 2019. PeerJ. 2019. PMID: 31592339 Free PMC article.
-
Molecular and clinical insights from studies of calcium-sensing receptor mutations.J Mol Endocrinol. 2019 Aug;63(2):R1-R16. doi: 10.1530/JME-19-0104. J Mol Endocrinol. 2019. PMID: 31189130 Review.
-
Insights into calcium-sensing receptor trafficking and biased signalling by studies of calcium homeostasis.J Mol Endocrinol. 2018 Jul;61(1):R1-R12. doi: 10.1530/JME-18-0049. Epub 2018 Mar 29. J Mol Endocrinol. 2018. PMID: 29599414 Review.
Cited by
-
Identification of acetylcholinesterase inhibitors from traditional medicinal plants for Alzheimer's disease using in silico and machine learning approaches.RSC Adv. 2024 Oct 31;14(47):34620-34636. doi: 10.1039/d4ra05073h. eCollection 2024 Oct 29. RSC Adv. 2024. PMID: 39483377 Free PMC article.
-
3D-QSAR and Molecular Dynamics Study of Isoxazole Derivatives to Identify the Structural Requirements for Farnesoid X Receptor (FXR) Agonists.Molecules. 2024 Mar 8;29(6):1210. doi: 10.3390/molecules29061210. Molecules. 2024. PMID: 38542850 Free PMC article.
-
Profiling and cheminformatics bioprospection of curcurbitacin I and momordin Ic from Momordica balsamina on α-amylase and α-glucosidase.J Enzyme Inhib Med Chem. 2025 Dec;40(1):2492706. doi: 10.1080/14756366.2025.2492706. Epub 2025 Apr 29. J Enzyme Inhib Med Chem. 2025. PMID: 40302171 Free PMC article.
-
The Italian registry of families at risk for pancreatic cancer (IRFARPC): implementation and evolution of a national program for pancreatic cancer surveillance in high-risk individuals.Fam Cancer. 2024 Aug;23(3):373-382. doi: 10.1007/s10689-024-00366-3. Epub 2024 Mar 16. Fam Cancer. 2024. PMID: 38493228 Review.
-
High-Throughput Molecular Modeling and Evaluation of the Anti-Inflammatory Potential of Açaí Constituents against NLRP3 Inflammasome.Int J Mol Sci. 2024 Jul 25;25(15):8112. doi: 10.3390/ijms25158112. Int J Mol Sci. 2024. PMID: 39125681 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

