Absolute quantification and characterization of oxylipins in lupus nephritis and systemic lupus erythematosus
- PMID: 36275708
- PMCID: PMC9582137
- DOI: 10.3389/fimmu.2022.964901
Absolute quantification and characterization of oxylipins in lupus nephritis and systemic lupus erythematosus
Abstract
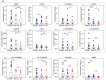
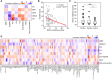
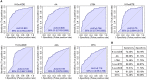
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease with multi-organ inflammation and defect, which is linked to many molecule mediators. Oxylipins as a class of lipid mediator have not been broadly investigated in SLE. Here, we applied targeted mass spectrometry analysis to screen the alteration of oxylipins in serum of 98 SLE patients and 106 healthy controls. The correlation of oxylipins to lupus nephritis (LN) and SLE disease activity, and the biomarkers for SLE classification, were analyzed. Among 128 oxylipins analyzed, 92 were absolutely quantified and 26 were significantly changed. They were mainly generated from the metabolism of several polyunsaturated fatty acids, including arachidonic acid (AA), linoleic acid (LA), docosahexanoic acid (DHA), eicosapentanoic acid (EPA) and dihomo-γ-linolenic acid (DGLA). Several oxylipins, especially those produced from AA, showed different abundance between patients with and without lupus nephritis (LN). The DGLA metabolic activity and DGLA generated PGE1, were significantly associated with SLE disease activity. Random forest-based machine learning identified a 5-oxylipin combination as potential biomarker for SLE classification with high accuracy. Seven individual oxylipin biomarkers were also identified with good performance in distinguishing SLE patients from healthy controls (individual AUC > 0.7). Interestingly, the biomarkers for differentiating SLE patients from healthy controls are distinct from the oxylipins differentially expressed in LN patients vs. non-LN patients. This study provides possibilities for the understanding of SLE characteristics and the development of new tools for SLE classification.
Keywords: SLEDAI; lupus nephritis; oxylipin; polyunsaturated fatty acids; systemic lupus erythematosus.
Copyright © 2022 He, Ma, Tang, Zhong, Yuan, Zheng, Zeng, Chen, Liu, Hong, Dai, Yin and Dai.
Conflict of interest statement
SZ and XY are employed by the company Shanghai Biotree Biomedical Biotechnology Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

