Multilocus sequence typing of Giardia duodenalis genotypes circulating in humans in a major metropolitan area
- PMID: 36275791
- PMCID: PMC9581142
- DOI: 10.3389/fmed.2022.976956
Multilocus sequence typing of Giardia duodenalis genotypes circulating in humans in a major metropolitan area
Abstract
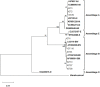
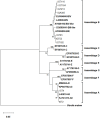
Giardia duodenalis is an intestinal protozoan parasite of humans and animal hosts and comprises eight microscopically indistinguishable molecularly-diverse lineages designated as assemblages A-H. Assemblages A and B are the primary sources of infections in humans and a wide range of mammals. Here, we identified assemblages, and inter-/intra-assemblage genetic diversity of human G. duodenalis isolates based on the multilocus sequence typing of the triosephosphate isomerase (tpi), β -giardin (bg), and glutamate dehydrogenase (gdh) loci. Multilocus sequence analysis of 62 microscopically-positive G. duodenalis fecal samples identified 26 (41.9%), 27 (43.5%), and nine (14.5%) isolates belonging to assemblages A, B, and discordant assemblages, respectively. The tpi locus assemblage-specific primers identified dual infections with A and B assemblages (45.2%). The sequence analysis of multiple alignments and phylogenetic analysis showed low genetic polymorphism in assemblage A isolates, classified as sub-assemblage AII at three loci, subtype A2 at tpi and gdh loci, and subtype A2 or A3 at bg locus. High genetic variations were found in assemblage B isolates with 14, 15, and 23 nucleotide patterns at tpi, bg, and gdh loci, respectively. Further concatenated sequence analysis revealed four multilocus genotypes (MLG) in 24 assemblages A isolates, two previously-identified (AII-1 and AII-5), with one novel multilocus genotype. However, the high genetic variations observed in assemblage B isolates among and within the three genetic loci prevented the definitive designation of specific MLGs for these isolates. Multilocus sequence typing may provide new insight into the genetic diversity of G. duodenalis isolates in Tehran, suggesting that humans are likely a potential source of G. duodenalis infection. Further host-specific experimental transmission studies are warranted to elucidate the modes of transmission within multiple host populations.
Keywords: Giardia duodenalis; Iran; Tehran; glutamate dehydrogenase (gdh); multilocus sequence typing; triosephosphate isomerase (tpi); β-giardin (bg).
Copyright © 2022 Hashemi-Hafshejani, Meamar, Moradi, Hemmati, Solaymani-Mohammadi and Razmjou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
High genetic diversity of Giardia duodenalis assemblage E in pre-weaned dairy calves in Shanghai, China, revealed by multilocus genotyping.Parasitol Res. 2017 Aug;116(8):2101-2110. doi: 10.1007/s00436-017-5509-8. Epub 2017 May 26. Parasitol Res. 2017. PMID: 28550644
-
Genetic Diversity and Prevalence of Giardia duodenalis in Qatar.Front Cell Infect Microbiol. 2021 May 4;11:652946. doi: 10.3389/fcimb.2021.652946. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34017691 Free PMC article.
-
Occurrence and multilocus genotyping of Giardia duodenalis in black-boned sheep and goats in southwestern China.Parasit Vectors. 2019 Mar 13;12(1):102. doi: 10.1186/s13071-019-3367-1. Parasit Vectors. 2019. PMID: 30867035 Free PMC article.
-
Multilocus genotyping of Giardia duodenalis in Bactrian camels (Camelus bactrianus) in China.Parasitol Res. 2020 Nov;119(11):3873-3880. doi: 10.1007/s00436-020-06905-y. Epub 2020 Oct 2. Parasitol Res. 2020. PMID: 33006040
-
Giardia duodenalis Infections in Humans and Other Animals in China.Front Microbiol. 2017 Oct 13;8:2004. doi: 10.3389/fmicb.2017.02004. eCollection 2017. Front Microbiol. 2017. PMID: 29081771 Free PMC article. Review.
Cited by
-
Molecular identification of Cryptosporidium, Giardia, and Blastocystis from stray and household cats and cat owners in Tehran, Iran.Sci Rep. 2023 Jan 27;13(1):1554. doi: 10.1038/s41598-023-28768-w. Sci Rep. 2023. PMID: 36707690 Free PMC article.
-
A shotgun metagenomic analysis of the fecal microbiome in humans infected with Giardia duodenalis.Parasit Vectors. 2023 Jul 18;16(1):239. doi: 10.1186/s13071-023-05821-1. Parasit Vectors. 2023. PMID: 37464386 Free PMC article.
-
Molecular characterization and risk analysis of Giardia duodenalis assemblages in corticosteroid-treated and non-treated patients in Ismailia, Arab Republic of Egypt.Gut Pathog. 2024 Dec 13;16(1):74. doi: 10.1186/s13099-024-00668-y. Gut Pathog. 2024. PMID: 39673061 Free PMC article.
-
Molecular epidemiology and multilocus genotyping of Giardia duodenalis in individuals attending major public hospitals in Shiraz, southwestern Iran: A public health concern.Parasite Epidemiol Control. 2024 Apr 29;25:e00354. doi: 10.1016/j.parepi.2024.e00354. eCollection 2024 May. Parasite Epidemiol Control. 2024. PMID: 38711926 Free PMC article.
-
Molecular Detection and Epidemiology of Potentially Zoonotic Cryptosporidium spp. and Giardia duodenalis in Wild Boar (Sus scrofa) from Eastern Spain.Animals (Basel). 2023 Aug 3;13(15):2501. doi: 10.3390/ani13152501. Animals (Basel). 2023. PMID: 37570308 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

