Integrated computational approach towards identification of HSPG and ACE2 mimicking moieties for SARS-CoV-2 inhibition
- PMID: 36276265
- PMCID: PMC9578758
- DOI: 10.1016/j.molliq.2022.120566
Integrated computational approach towards identification of HSPG and ACE2 mimicking moieties for SARS-CoV-2 inhibition
Abstract
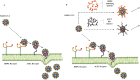
A key step to inhibit severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is to prevent the entry of the virus into the host cells. The receptor-binding domains (RBDs) of spike proteins of SARS-CoV and other human coronaviruses utilize heparan sulfate proteoglycans (HSPGs) as the primary receptors for their accumulation on the cell surface and then scan for binding to the main entry receptor angiotensin-converting enzyme 2 (ACE2). SARS-CoV and SARS-CoV-2 share structurally similar RBDs and therefore, it is possible that SARS-COV-2 primarily binds to HSPGs followed by binding to the ACE2 receptors. A promising strategy to inhibit virus infection is to circulate exogenous bioactive moieties structurally mimicking cellular HSPG and ACE2 which act as decoy receptors binding to SARS-CoV-2 and competitively inhibit virus entry to the host cells mediated by cellular-bound HSPG and ACE2. Using a molecular docking tool, we identified carboxymethyl benzyl amide sulfonate (CMBS) and polyanetholesulfonic acid (PAS) as the suitable HSPG mimicking ligands, and Paenibacillus sp. B38-derived carboxypeptidase (B38-CAP) and Bacillus subtilis-derived carboxypeptidase (BS-CAP) as the potential ACE2-like enzymes having a strong binding affinity to the spike proteins as that of cellular HSPG and ACE2. Further, the binding stability and compactness of these moieties with SARS-CoV-2 were analyzed through molecular dynamics (MD) simulations, and the results indicated that these moieties form well-stable complexes with the RBD of spike proteins. The identified moieties could be conjugated to the surfaces of non-toxic nanoparticles to provide multiple interactions to efficiently shield SARS-CoV-2, and inhibit viral entry to the host cells.
Keywords: ACE2; HSPG; Inhibition; Mimicking moieties; Molecular docking; Molecular dynamics simulations; SARS-CoV-2.
© 2022 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures






Similar articles
-
Screening of inhibitors against SARS-CoV-2 spike protein and their capability to block the viral entry mechanism: A viroinformatics study.Saudi J Biol Sci. 2021 Jun;28(6):3262-3269. doi: 10.1016/j.sjbs.2021.02.066. Epub 2021 Feb 26. Saudi J Biol Sci. 2021. PMID: 33654454 Free PMC article.
-
Withanone from Withania somnifera Attenuates SARS-CoV-2 RBD and Host ACE2 Interactions to Rescue Spike Protein Induced Pathologies in Humanized Zebrafish Model.Drug Des Devel Ther. 2021 Mar 11;15:1111-1133. doi: 10.2147/DDDT.S292805. eCollection 2021. Drug Des Devel Ther. 2021. PMID: 33737804 Free PMC article.
-
Evolutionary Arms Race between Virus and Host Drives Genetic Diversity in Bat Severe Acute Respiratory Syndrome-Related Coronavirus Spike Genes.J Virol. 2020 Sep 29;94(20):e00902-20. doi: 10.1128/JVI.00902-20. Print 2020 Sep 29. J Virol. 2020. PMID: 32699095 Free PMC article.
-
Therapeutic development targeting host heparan sulfate proteoglycan in SARS-CoV-2 infection.Front Med (Lausanne). 2024 Mar 27;11:1364657. doi: 10.3389/fmed.2024.1364657. eCollection 2024. Front Med (Lausanne). 2024. PMID: 38618194 Free PMC article. Review.
-
Inhibition of S-protein RBD and hACE2 Interaction for Control of SARSCoV- 2 Infection (COVID-19).Mini Rev Med Chem. 2021;21(6):689-703. doi: 10.2174/1389557520666201117111259. Mini Rev Med Chem. 2021. PMID: 33208074 Review.
Cited by
-
Interaction of SARS-CoV-2 and SARS-CoV-2 vaccines with renin angiotensin aldosterone system, clinical outcomes, and angiotensin (1-7) as a physiological treatment recommendation: hypothesis and theory article.Front Med (Lausanne). 2025 Jul 10;12:1612442. doi: 10.3389/fmed.2025.1612442. eCollection 2025. Front Med (Lausanne). 2025. PMID: 40708634 Free PMC article.
References
-
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B.C., features of patients infected with, novel coronavirus in Wuhan, China. Lancet. 2019;395(2020):497–506. doi: 10.1016/S0140-6736(20)30183-5. - DOI - PMC - PubMed
-
- WHO Coronavirus (COVID-19) Dashboard, https://covid19.who.int/.
LinkOut - more resources
Full Text Sources
Miscellaneous
