The lncRNA KIF9-AS1 Accelerates Hepatocellular Carcinoma Growth by Recruiting DNMT1 to Promote RAI2 DNA Methylation
- PMID: 36276278
- PMCID: PMC9584731
- DOI: 10.1155/2022/3888798
The lncRNA KIF9-AS1 Accelerates Hepatocellular Carcinoma Growth by Recruiting DNMT1 to Promote RAI2 DNA Methylation
Abstract
Background: Hepatocellular carcinoma (HCC) is a very common malignant tumor. Long noncoding RNAs (lncRNAs) enable discoveries of new therapeutic tumor targets. We aimed to study the role and potential regulatory mechanisms of the lncRNA KIF9-AS1 in HCC.
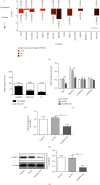
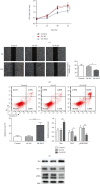
Methods: CCK-8, scratch assay, and flow cytometry were used to detect cell proliferation, migration, and apoptosis, respectively. Bax, Bcl-2, ERK, and pERK expression were measured by western blotting. StarBase predicted KIF9-AS1 expression in HCC and paracancerous tissues. RPISeq predicted the interaction score of KIF9-AS1 and DNMT1, and MethyPrimer revealed the CpG island distribution in the RAI2 promoter. MSP was performed to measure RAI2 methylation. RIP and ChIP were performed to examine lncRNA KIF9-AS1, DNMT1, and RAI2 interactions. Finally, the effect of KIF9-AS1 knockdown on HCC was verified with nude mice.
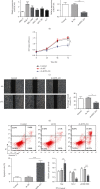
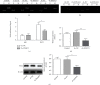
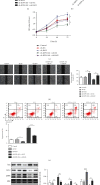
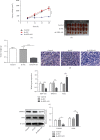
Results: We found that KIF9-AS1 expression was increased in HCC tissues. KIF9-AS1 knockdown inhibited the proliferation and migration, and facilitated the apoptosis of HCC cells. lncRNA KIF9-AS1-mediated RAI2 expression led to DNMT1 recruitment and regulated RAI2 DNA methylation. RAI2 overexpression inhibited the proliferation and migration and promoted the apoptosis of HCC cells. KIF9-AS1 knockdown inhibited subcutaneous tumor formation in vivo.
Conclusion: This study shows that KIF9-AS1 accelerates HCC growth by inducing DNMT1 promotion of RAI2 DNA methylation.
Copyright © 2022 Yong Yu et al.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

