Identifying the E2F3-MEX3A-KLF4 signaling axis that sustains cancer cells in undifferentiated and proliferative state
- PMID: 36276637
- PMCID: PMC9576623
- DOI: 10.7150/thno.76619
Identifying the E2F3-MEX3A-KLF4 signaling axis that sustains cancer cells in undifferentiated and proliferative state
Abstract
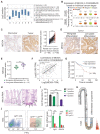
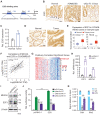
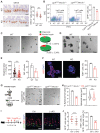
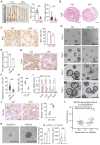
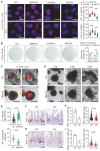
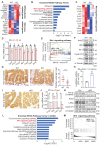
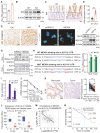
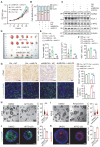
Rationale: Dysregulation of signaling that governs self-renewal and differentiation of intestinal stem cells (ISCs) is a major cause of colorectal cancer (CRC) initiation and progression. Methods: qRT-PCR, western blotting, in situ hybridization, immunohistochemistry and immunofluorescence assays were used to detect the expression levels of MEX3A, KLF4 and E2F3 in CRC tissues. The biological functions of MEX3A were studied using Mex3a knockout (KO) and intestinal epithelium specific conditional knockout (cKO) mice, AOM-DSS mouse colorectal tumor model, Apc floxed mouse tumor model and intestinal and tumor organoids. Transcriptomic RNA sequencing (RNA-seq), RNA crosslinking immunoprecipitation (CLIP) and luciferase reporter assays were performed to explore the molecular mechanisms of MEX3A. Results: RNA-binding protein MEX3A, a specific ISC marker gene, becomes ectopically upregulated upon CRC and its levels negatively correlate with patient survival prognosis. MEX3A functions as an oncoprotein that retains cancer cells in undifferentiated and proliferative status and it enhances their radioresistance to DNA damage. Mechanistically, a rate limiting factor of cellular proliferation E2F3 induces MEX3A, which in turn activates WNT pathway by directly suppressing expression of its pro-differentiation transcription factor KLF4. Knockdown of MEX3A with siRNA or addition of KLF4 agonist significantly suppressed tumor growth both by increasing differentiation status of cancer cells and by suppressing their proliferation. Conclusions: It identifies E2F3-MEX3A-KLF4 axis as an essential coordinator of cancer stem cell self-renewal and differentiation, representing a potent new druggable target for cancer differentiation therapy.
Keywords: Cancer stemness; Colorectal cancer; KLF4; MEX3A; Radio-resistance.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures
Similar articles
-
MEX3A promotes colorectal cancer migration, invasion and EMT via regulating the Wnt/β-catenin signaling pathway.J Cancer Res Clin Oncol. 2024 Jun 25;150(6):319. doi: 10.1007/s00432-024-05845-9. J Cancer Res Clin Oncol. 2024. PMID: 38914858 Free PMC article.
-
miR-503 inhibits cell proliferation and induces apoptosis in colorectal cancer cells by targeting E2F3.Int J Clin Exp Pathol. 2015 Oct 1;8(10):12853-60. eCollection 2015. Int J Clin Exp Pathol. 2015. PMID: 26722476 Free PMC article.
-
In Colorectal Cancer Cells With Mutant KRAS, SLC25A22-Mediated Glutaminolysis Reduces DNA Demethylation to Increase WNT Signaling, Stemness, and Drug Resistance.Gastroenterology. 2020 Dec;159(6):2163-2180.e6. doi: 10.1053/j.gastro.2020.08.016. Epub 2020 Aug 16. Gastroenterology. 2020. PMID: 32814111
-
SMYD2 epigenetically activates MEX3A and suppresses CDX2 in colorectal cancer cells to augment cancer growth.Clin Exp Pharmacol Physiol. 2022 Sep;49(9):959-969. doi: 10.1111/1440-1681.13679. Epub 2022 Jun 18. Clin Exp Pharmacol Physiol. 2022. PMID: 35637161
-
Oncogenic Potential of the Dual-Function Protein MEX3A.Biology (Basel). 2021 May 7;10(5):415. doi: 10.3390/biology10050415. Biology (Basel). 2021. PMID: 34067172 Free PMC article. Review.
Cited by
-
Homozygous editing of multiple genes for accelerated generation of xenotransplantation pigs.Genome Res. 2025 May 2;35(5):1167-1178. doi: 10.1101/gr.279709.124. Genome Res. 2025. PMID: 40044221 Free PMC article.
-
KLF4 Induces Colorectal Cancer by Promoting EMT via STAT3 Activation.Dig Dis Sci. 2024 Aug;69(8):2841-2855. doi: 10.1007/s10620-024-08473-y. Epub 2024 May 30. Dig Dis Sci. 2024. PMID: 38816600
-
Murine models of colorectal cancer: the azoxymethane (AOM)/dextran sulfate sodium (DSS) model of colitis-associated cancer.PeerJ. 2023 Oct 31;11:e16159. doi: 10.7717/peerj.16159. eCollection 2023. PeerJ. 2023. PMID: 37927787 Free PMC article. Review.
-
m6A Reader PRRC2A Promotes Colorectal Cancer Progression via CK1ε-Mediated Activation of WNT and YAP Signaling Pathways.Adv Sci (Weinh). 2025 Jan;12(3):e2406935. doi: 10.1002/advs.202406935. Epub 2024 Nov 24. Adv Sci (Weinh). 2025. PMID: 39582289 Free PMC article.
-
Dexamethasone-tamoxifen combination exerts synergistic therapeutic effects in tamoxifen-resistance breast cancer cells.Biosci Rep. 2024 Jul 31;44(7):BSR20240367. doi: 10.1042/BSR20240367. Biosci Rep. 2024. PMID: 38864530 Free PMC article.
References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. - PubMed
-
- Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–68. - PubMed
-
- van Es JH, Clevers H. Notch and Wnt inhibitors as potential new drugs for intestinal neoplastic disease. Trends Mol Med. 2005;11:496–502. - PubMed
-
- Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M. et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–11. - PubMed
-
- Zhang Y, Que J. BMP signaling in development, stem cells, and diseases of the gastrointestinal tract. Annu Rev of Physiol. 2020;82:251–73. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

