Addressing antimicrobial resistance with the IDentif.AI platform: Rapidly optimizing clinically actionable combination therapy regimens against nontuberculous mycobacteria
- PMID: 36276648
- PMCID: PMC9576615
- DOI: 10.7150/thno.73078
Addressing antimicrobial resistance with the IDentif.AI platform: Rapidly optimizing clinically actionable combination therapy regimens against nontuberculous mycobacteria
Abstract
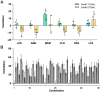
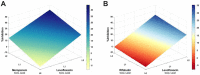
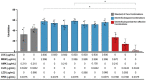
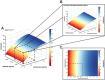
Background: Current standard of care (SOC) regimens against nontuberculous mycobacteria (NTM) usually result in unsatisfactory therapeutic responses, primarily due to multi-drug resistance and antibiotic susceptibility-guided therapies. In the midst of rising incidences in NTM infections, strategies to develop NTM-specific treatments have been explored and validated. Methods: To provide an alternative approach to address NTM-specific treatment, IDentif.AI was harnessed to rapidly optimize and design effective combination therapy regimens against Mycobacterium abscessus (M. abscessus), the highly resistant and rapid growth species of NTM. IDentif.AI interrogated the drug interaction space from a pool of 6 antibiotics, and pinpointed multiple clinically actionable drug combinations. IDentif.AI-pinpointed actionable combinations were experimentally validated and their interactions were assessed using Bliss independence model and diagonal measurement of n-way drug interactions. Results: Notably, IDentfi.AI-designed 3- and 4-drug combinations demonstrated greater %Inhibition efficacy than the SOC regimens. The platform also pinpointed two unique drug interactions (Levofloxacin (LVX)/Rifabutin (RFB) and LVX/Meropenem (MEM)) that may serve as the backbone of potential 3- and 4-drug combinations like LVX/MEM/RFB, which exhibited 58.33±4.99 %Inhibition efficacy against M. abscessus. Further analysis of LVX/RFB via Bliss independence model pointed to dose-dependent synergistic interactions in clinically actionable concentrations. Conclusions: IDentif.AI-designed combinations may provide alternative regimen options to current SOC combinations that are often administered with Amikacin, which has been known to induce ototoxicity in patients. Furthermore, IDentif.AI pinpointed 2-drug interactions may also serve as the backbone for the development of other effective 3- and 4-drug combination therapies. The findings in this study suggest that this platform may contribute to NTM-specific drug development.
Keywords: Mycobacterium abscessus; Artificial Intelligence; Combination Therapy; Infectious Disease; Nontuberculous Mycobacteria.
© The author(s).
Conflict of interest statement
Competing Interests: A.B., E.K.-H.C., and D.H. are co-inventors of previously filed pending patents on artificial intelligence-based therapy development. E.K.-H.C. and D.H. are co-founders and shareholders of KYAN Therapeutics, which is commercializing intellectual property pertaining to AI-based personalized medicine.
Figures

Similar articles
-
Clofazimine Prevents the Regrowth of Mycobacterium abscessus and Mycobacterium avium Type Strains Exposed to Amikacin and Clarithromycin.Antimicrob Agents Chemother. 2015 Dec 7;60(2):1097-105. doi: 10.1128/AAC.02615-15. Print 2016 Feb. Antimicrob Agents Chemother. 2015. PMID: 26643335 Free PMC article.
-
Antimicrobial susceptibility and minimum inhibitory concentration distribution of common clinically relevant non-tuberculous mycobacterial isolates from the respiratory tract.Ann Med. 2022 Dec;54(1):2500-2510. doi: 10.1080/07853890.2022.2121984. Ann Med. 2022. PMID: 36120867 Free PMC article.
-
Nontuberculous mycobacterial ocular infections--comparing the clinical and microbiological characteristics between Mycobacterium abscessus and Mycobacterium massiliense.PLoS One. 2015 Jan 12;10(1):e0116236. doi: 10.1371/journal.pone.0116236. eCollection 2015. PLoS One. 2015. PMID: 25581038 Free PMC article.
-
Resistance mechanisms and drug susceptibility testing of nontuberculous mycobacteria.Drug Resist Updat. 2012 Jun;15(3):149-61. doi: 10.1016/j.drup.2012.04.001. Epub 2012 Apr 21. Drug Resist Updat. 2012. PMID: 22525524 Review.
-
Recent advances in molecular diagnostics and understanding mechanisms of drug resistance in nontuberculous mycobacterial diseases.Infect Genet Evol. 2019 Aug;72:169-182. doi: 10.1016/j.meegid.2018.10.003. Epub 2018 Oct 11. Infect Genet Evol. 2019. PMID: 30315892 Review.
Cited by
-
Determining sex differences in aortic valve myofibroblast responses to drug combinations identified using a digital medicine platform.Sci Adv. 2025 Jun 6;11(23):eadu2695. doi: 10.1126/sciadv.adu2695. Epub 2025 Jun 6. Sci Adv. 2025. PMID: 40479052 Free PMC article.
-
Pharmacokinetics-Pharmacodynamics Modeling for Evaluating Drug-Drug Interactions in Polypharmacy: Development and Challenges.Clin Pharmacokinet. 2024 Jul;63(7):919-944. doi: 10.1007/s40262-024-01391-2. Epub 2024 Jun 18. Clin Pharmacokinet. 2024. PMID: 38888813 Review.
-
N-of-1 medicine.Singapore Med J. 2024 Mar 1;65(3):167-175. doi: 10.4103/singaporemedj.SMJ-2023-243. Epub 2024 Mar 26. Singapore Med J. 2024. PMID: 38527301 Free PMC article.
-
Flash optimization of drug combinations for Acinetobacter baumannii with IDentif.AI-AMR.NPJ Antimicrob Resist. 2025 Feb 21;3(1):12. doi: 10.1038/s44259-025-00079-2. NPJ Antimicrob Resist. 2025. PMID: 39984645 Free PMC article.
-
Deep Learning and Drug Discovery for Healthy Aging.ACS Cent Sci. 2023 Oct 17;9(10):1860-1863. doi: 10.1021/acscentsci.3c01212. eCollection 2023 Oct 25. ACS Cent Sci. 2023. PMID: 37901176 Free PMC article. No abstract available.
References
-
- Ahmed I, Tiberi S, Farooqi J, Jabeen K, Yeboah-Manu D, Migliori GB. et al. Non-tuberculous mycobacterial infections—A neglected and emerging problem. Int J Infect Dis. 2020;92:S46–S50. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

