The Comparison of Metagenomic Next-Generation Sequencing with Conventional Microbiological Tests for Identification of Pathogens and Antibiotic Resistance Genes in Infectious Diseases
- PMID: 36277249
- PMCID: PMC9586124
- DOI: 10.2147/IDR.S370964
The Comparison of Metagenomic Next-Generation Sequencing with Conventional Microbiological Tests for Identification of Pathogens and Antibiotic Resistance Genes in Infectious Diseases
Abstract
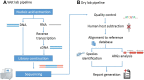
Background: Metagenomic next-generation sequencing (mNGS) has been widely studied, due to its ability of detecting all the microbial genetic information unbiasedly in a sample at one time and not relying on traditional culture. However, the application of mNGS in the diagnosis of clinical pathogens remains challenging.
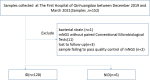
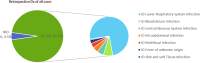
Methods: From December 2019 to March 2021, 134 specimens including Broncho alveolar lavage fluid (BAFL), blood, sputum, cerebrospinal fluid (CSF), bile, pleural fluid, pus, were continuously collected in The First Hospital of Qinhuangdao, and their retrospective diagnoses were classified into infectious disease (128, 95.5%) and noninfectious disease (6, 4.5%). The pathogen-detection performance of mNGS was compared with conventional microbiological tests (CMT) and culture method. In addition, the antibiotic resistance genes (ARGs) and evolutionary relationship of common drug-resistant A. baumannii were also analyzed.
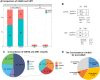
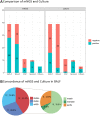
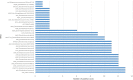
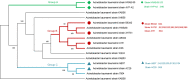
Results: Compared with CMT and culture methods, mNGS showed higher sensitivity in pathogen detection (74.2% vs 57.8%; P < 0.001 and 66.3% vs 31.7%; P < 0.001, respectively). Importantly, for cases that mNGS-positive only, 18 (35%) cases result in diagnosis modification, and 7 (23%) cases confirmed the clinical diagnosis. In 17 cases that A. baumannii were both detected in mNGS and culture, ade genes were the most frequently detected ARGs (from 13 cases), followed by sul2 and APH(3")-Ib (both from 12 cases). High consistency was observed among these ARGs and the related phenotype (100% for ade genes, 91.6% for sul2 and APH(3")-Ib). A. baumannii strains were classified into three groups, and most were well-clustered. It suggested those strains may be the epidemic strains.
Conclusion: In our study, mNGS had a higher sensitivity than CMT and culture method. And the result of ARGs frequency and cluster analysis of A. baumannii was of great significance to the anti-infective therapy.
Keywords: antibiotic resistance genes; conventional microbiological tests; infection; metagenomic next-generation sequencing; sensitivity.
© 2022 Lu et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Clinical Efficiency of Metagenomic Next-Generation Sequencing in Sputum for Pathogen Detection of Patients with Pneumonia According to Disease Severity and Host Immune Status.Infect Drug Resist. 2023 Sep 6;16:5869-5885. doi: 10.2147/IDR.S419892. eCollection 2023. Infect Drug Resist. 2023. PMID: 37700802 Free PMC article.
-
The diagnostic value of metagenomic next⁃generation sequencing in infectious diseases.BMC Infect Dis. 2021 Jan 13;21(1):62. doi: 10.1186/s12879-020-05746-5. BMC Infect Dis. 2021. PMID: 33435894 Free PMC article.
-
Clinical utility of metagenomic next-generation sequencing in pathogen detection for lower respiratory tract infections and impact on clinical outcomes in southernmost China.Front Cell Infect Microbiol. 2023 Dec 8;13:1271952. doi: 10.3389/fcimb.2023.1271952. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 38145053 Free PMC article.
-
The clinical application of metagenomic next-generation sequencing for detecting pathogens in bronchoalveolar lavage fluid: case reports and literature review.Expert Rev Mol Diagn. 2022 May;22(5):575-582. doi: 10.1080/14737159.2022.2071607. Epub 2022 May 2. Expert Rev Mol Diagn. 2022. PMID: 35473493 Review.
-
Metagenomics next-generation sequencing tests take the stage in the diagnosis of lower respiratory tract infections.J Adv Res. 2021 Sep 29;38:201-212. doi: 10.1016/j.jare.2021.09.012. eCollection 2022 May. J Adv Res. 2021. PMID: 35572406 Free PMC article. Review.
Cited by
-
Diagnostic Role of Metagenomic Next-Generation Sequencing in Tubercular Orthopedic Implant-Associated Infection.Infect Drug Resist. 2024 May 17;17:1951-1960. doi: 10.2147/IDR.S441940. eCollection 2024. Infect Drug Resist. 2024. PMID: 38774035 Free PMC article.
-
Comparison of metagenomic next-generation sequencing and conventional culture for the diagnostic performance in febrile patients with suspected infections.BMC Infect Dis. 2024 Mar 26;24(1):350. doi: 10.1186/s12879-024-09236-w. BMC Infect Dis. 2024. PMID: 38532348 Free PMC article.
-
Microbiological diagnostic performance of metagenomic next-generation sequencing compared with conventional culture for patients with community-acquired pneumonia.Front Cell Infect Microbiol. 2023 Mar 16;13:1136588. doi: 10.3389/fcimb.2023.1136588. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37009509 Free PMC article.
-
Clinical Efficiency of Metagenomic Next-Generation Sequencing in Sputum for Pathogen Detection of Patients with Pneumonia According to Disease Severity and Host Immune Status.Infect Drug Resist. 2023 Sep 6;16:5869-5885. doi: 10.2147/IDR.S419892. eCollection 2023. Infect Drug Resist. 2023. PMID: 37700802 Free PMC article.
-
Clinical Efficacy and Diagnostic Value of Metagenomic Next-Generation Sequencing for Pathogen Detection in Patients with Suspected Infectious Diseases: A Retrospective Study from a Large Tertiary Hospital.Infect Drug Resist. 2023 Mar 29;16:1815-1828. doi: 10.2147/IDR.S401707. eCollection 2023. Infect Drug Resist. 2023. PMID: 37016633 Free PMC article.
References
-
- Troeger C, Blacker B, Khalil IA; Collaborators GBDLRI. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect Dis. 2018;18(11):1191–1210. doi:10.1016/S1473-3099(18)30310-4 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

