Muscle transcriptomic circuits linked to periarticular physiology in end-stage osteoarthritis
- PMID: 36278270
- PMCID: PMC9762959
- DOI: 10.1152/physiolgenomics.00092.2022
Muscle transcriptomic circuits linked to periarticular physiology in end-stage osteoarthritis
Abstract
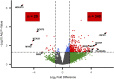
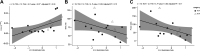
The ability of individuals with end-stage osteoarthritis (OA) to functionally recover from total joint arthroplasty is highly inconsistent. The molecular mechanisms driving this heterogeneity have yet to be elucidated. Furthermore, OA disproportionately impacts females, suggesting a need for identifying female-specific therapeutic targets. We profiled the skeletal muscle transcriptome in females with end-stage OA (n = 20) undergoing total knee or hip arthroplasty using RNA-Seq. Single-gene differential expression (DE) analyses tested for DE genes between skeletal muscle overlaying the surgical (SX) joint and muscle from the contralateral (CTRL) leg. Network analyses were performed using Pathway-Level Information ExtractoR (PLIER) to summarize genes into latent variables (LVs), i.e., gene circuits, and link them to biological pathways. LV differences in SX versus CTRL muscle and across sources of muscle tissue (vastus medialis, vastus lateralis, or tensor fascia latae) were determined with ANOVA. Linear models tested for associations between LVs and muscle phenotype on the SX side (inflammation, function, and integrity). DE analysis revealed 360 DE genes (|Log2 fold-difference| ≥ 1, FDR ≤ 0.05) between the SX and CTRL limbs, many associated with inflammation and lipid metabolism. PLIER analyses revealed circuits associated with protein degradation and fibro-adipogenic cell gene expression. Muscle inflammation and function were linked to an LV associated with endothelial cell gene expression highlighting a potential regulatory role of endothelial cells within skeletal muscle. These findings may provide insight into potential therapeutic targets to improve OA rehabilitation before and/or following total joint replacement.
Keywords: osteoarthritis; skeletal muscle; transcriptomics.
Conflict of interest statement
J.A.S. has received consultant fees from Schipher, Crealta/Horizon, Medisys, Fidia, PK Med, Two laboratories Inc., Adept Field Solutions, Clinical Care options, Clearview healthcare partners, Putnam associates, Focus forward, Navigant consulting, Spherix, MedIQ, Jupiter Life Science, UBM LLC, Trio Health, Medscape, WebMD, and Practice Point communications; and the National Institutes of Health and the American College of Rheumatology. J.A.S. has received institutional research support from Zimmer Biomet Holdings. J.A.S. received food and beverage payments from Intuitive Surgical Inc./Philips Electronics North America. J.A.S. owns stock options in TPT Global Tech, Vaxart pharmaceuticals, Atyu biopharma, Adaptimmune Therapeutics, GeoVax Labs, Pieris Pharmaceuticals, Enzolytics Inc., Seres Therapeutics, Tonix Pharmaceuticals Holding Corp., and Charlotte’s Web Holdings, Inc. J.A.S. previously owned stock options in Amarin, Viking and Moderna pharmaceuticals. J.A.S. is on the speaker’s bureau of Simply Speaking. J.A.S. is a member of the executive of Outcomes Measures in Rheumatology (OMERACT), an organization that develops outcome measures in rheumatology and receives arms-length funding from 8 companies. J.A.S. serves on the FDA Arthritis Advisory Committee. J.A.S. is the chair of the Veterans Affairs Rheumatology Field Advisory Committee. J.A.S. is the editor and the Director of the University of Alabama at Birmingham (UAB) Cochrane Musculoskeletal Group Satellite Center on Network Meta-analysis. J.A.S. previously served as a member of the following committees: member, the American College of Rheumatology’s (ACR) Annual Meeting Planning Committee (AMPC) and Quality of Care Committees, the Chair of the ACR Meet-the-Professor, Workshop and Study Group Subcommittee and the co-chair of the ACR Criteria and Response Criteria subcommittee. None of the other authors has any conflicts of interest, financial or otherwise, to disclose.
Figures




References
-
- Safiri S, Kolahi A-A, Smith E, Hill C, Bettampadi D, Mansournia MA, Hoy D, Ashrafi-Asgarabad A, Sepidarkish M, Almasi-Hashiani A, Collins G, Kaufman J, Qorbani M, Moradi-Lakeh M, Woolf AD, Guillemin F, March L, Cross M. Global, regional and national burden of osteoarthritis 1990-2017: a systematic analysis of the Global Burden of Disease Study 2017. Ann Rheum Dis 79: 819–828, 2020. doi: 10.1136/annrheumdis-2019-216515. - DOI - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

