SIRT6 Mitigates Heart Failure With Preserved Ejection Fraction in Diabetes
- PMID: 36278398
- PMCID: PMC9669223
- DOI: 10.1161/CIRCRESAHA.121.318988
SIRT6 Mitigates Heart Failure With Preserved Ejection Fraction in Diabetes
Erratum in
-
Correction to: SIRT6 Mitigates Heart Failure With Preserved Ejection Fraction in Diabetes.Circ Res. 2023 Jul 21;133(3):e48. doi: 10.1161/RES.0000000000000622. Epub 2023 Jul 20. Circ Res. 2023. PMID: 37471488 No abstract available.
Abstract
Background: Heart failure with preserved ejection fraction (HFpEF) is a growing health problem without effective therapies. Epidemiological studies indicate that diabetes is a strong risk factor for HFpEF, and about 45% of patients with HFpEF are suffering from diabetes, yet the underlying mechanisms remain elusive.
Methods: Using a combination of echocardiography, hemodynamics, RNA-sequencing, molecular biology, in vitro and in vivo approaches, we investigated the roles of SIRT6 (sirtuin 6) in regulation of endothelial fatty acid (FA) transport and HFpEF in diabetes.
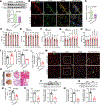
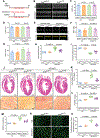
Results: We first observed that endothelial SIRT6 expression was markedly diminished in cardiac tissues from heart failure patients with diabetes. We then established an experimental mouse model of HFpEF in diabetes induced by a combination of the long-term high-fat diet feeding and a low-dose streptozocin challenge. We also generated a unique humanized SIRT6 transgenic mouse model, in which a single copy of human SIRT6 transgene was engineered at mouse Rosa26 locus and conditionally induced with the Cre-loxP technology. We found that genetically restoring endothelial SIRT6 expression in the diabetic mice ameliorated diastolic dysfunction concurrently with decreased cardiac lipid accumulation. SIRT6 gain- or loss-of-function studies showed that SIRT6 downregulated endothelial FA uptake. Mechanistically, SIRT6 suppressed endothelial expression of PPARγ through SIRT6-dependent deacetylation of histone H3 lysine 9 around PPARγ promoter region; and PPARγ reduction mediated SIRT6-dependent inhibition of endothelial FA uptake. Importantly, oral administration of small molecule SIRT6 activator MDL-800 to diabetic mice mitigated cardiac lipid accumulation and diastolic dysfunction.
Conclusions: The impairment of endothelial SIRT6 expression links diabetes to HFpEF through the alteration of FA transport across the endothelial barrier. Genetic and pharmacological strategies that restored endothelial SIRT6 function in mice with diabetes alleviated experimental HFpEF by limiting FA uptake and improving cardiac metabolism, thus warranting further clinical evaluation.
Keywords: HFpEF; PPARγ; diabetes; endothelial cells; fatty acid transport; heart failure; sirtuins.
Conflict of interest statement
Disclosures
A patent application related to this work has been filed. Z.G.J. is a founder of Bailey Pharmaceutical Technologies, Inc. No other competing interests.
Figures






References
-
- Shah SJ, Borlaug BA, Kitzman DW, McCulloch AD, Blaxall BC, Agarwal R, Chirinos JA, Collins S, Deo RC, Gladwin MT, et al. Research priorities for heart failure with preserved ejection fraction: National heart, lung, and blood institute working group summary. Circulation. 2020;141:1001–1026. doi: 10.1161/CIRCULATIONAHA.119.041886. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

