Proteomics-Based Identification of Dysregulated Proteins in Breast Cancer
- PMID: 36278695
- PMCID: PMC9590004
- DOI: 10.3390/proteomes10040035
Proteomics-Based Identification of Dysregulated Proteins in Breast Cancer
Abstract
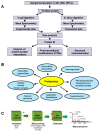
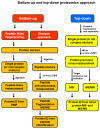
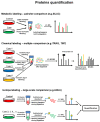
Immunohistochemistry (IHC) is still widely used as a morphology-based assay for in situ analysis of target proteins as specific tumor antigens. However, as a very heterogeneous collection of neoplastic diseases, breast cancer (BC) requires an accurate identification and characterization of larger panels of candidate biomarkers, beyond ER, PR, and HER2 proteins, for diagnosis and personalized treatment, without the limited availability of antibodies that are required to identify specific proteins. Top-down, middle-down, and bottom-up mass spectrometry (MS)-based proteomics approaches complement traditional histopathological tissue analysis to examine expression, modification, and interaction of hundreds to thousands of proteins simultaneously. In this review, we discuss the proteomics-based identification of dysregulated proteins in BC that are essential for the following issues: discovery and validation of new biomarkers by analysis of solid and liquid/non-invasive biopsies, cell lines, organoids and xenograft models; identification of panels of biomarkers for early detection and accurate discrimination between cancer, benign and normal tissues; identification of subtype-specific and stage-specific protein expression profiles in BC grading and measurement of disease progression; characterization of new subtypes of BC; characterization and quantitation of post-translational modifications (PTMs) and aberrant protein-protein interactions (PPI) involved in tumor development; characterization of the global remodeling of BC tissue homeostasis, diagnosis and prognostic information; and deciphering of molecular functions, biological processes and mechanisms through which the dysregulated proteins cause tumor initiation, invasion, and treatment resistance.
Keywords: breast cancer; dysregulated proteins; proteoforms; proteomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

