Imaging and Editing the Phospholipidome
- PMID: 36278840
- PMCID: PMC9703472
- DOI: 10.1021/acs.accounts.2c00510
Imaging and Editing the Phospholipidome
Abstract
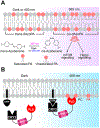
Membranes are multifunctional supramolecular assemblies that encapsulate our cells and the organelles within them. Glycerophospholipids are the most abundant component of membranes. They make up the majority of the lipid bilayer and play both structural and functional roles. Each organelle has a different phospholipid composition critical for its function that results from dynamic interplay and regulation of numerous lipid-metabolizing enzymes and lipid transporters. Because lipid structures and localizations are not directly genetically encoded, chemistry has much to offer to the world of lipid biology in the form of precision tools for visualizing lipid localization and abundance, manipulating lipid composition, and in general decoding the functions of lipids in cells.In this Account, we provide an overview of our recent efforts in this space focused on two overarching and complementary goals: imaging and editing the phospholipidome. On the imaging front, we have harnessed the power of bioorthogonal chemistry to develop fluorescent reporters of specific lipid pathways. Substantial efforts have centered on phospholipase D (PLD) signaling, which generates the humble lipid phosphatidic acid (PA) that acts variably as a biosynthetic intermediate and signaling agent. Though PLD is a hydrolase that generates PA from abundant phosphatidylcholine (PC) lipids, we have exploited its transphosphatidylation activity with exogenous clickable alcohols followed by bioorthogonal tagging to generate fluorescent lipid reporters of PLD signaling in a set of methods termed IMPACT.IMPACT and its variants have facilitated many biological discoveries. Using the rapid and fluorogenic tetrazine ligation, it has revealed the spatiotemporal dynamics of disease-relevant G protein-coupled receptor signaling and interorganelle lipid transport. IMPACT using diazirine photo-cross-linkers has enabled identification of lipid-protein interactions relevant to alcohol-related diseases. Varying the alcohol reporter can allow for organelle-selective labeling, and varying the bioorthogonal detection reagent can afford super-resolution lipid imaging via expansion microscopy. Combination of IMPACT with genome-wide CRISPR screening has revealed genes that regulate physiological PLD signaling.PLD enzymes themselves can also act as tools for precision editing of the phospholipid content of membranes. An optogenetic PLD for conditional blue-light-stimulated synthesis of PA on defined organelle compartments led to the discovery of the role of organelle-specific pools of PA in regulating oncogenic Hippo signaling. Directed enzyme evolution of PLD, enabled by IMPACT, has yielded highly active superPLDs with broad substrate tolerance and an ability to edit membrane phospholipid content and synthesize designer phospholipids in vitro. Finally, azobenzene-containing PA analogues represent an alternative, all-chemical strategy for light-mediated control of PA signaling.Collectively, the strategies described here summarize our progress to date in tackling the challenge of assigning precise functions to defined pools of phospholipids in cells. They also point to new challenges and directions for future study, including extension of imaging and membrane editing tools to other classes of lipids. We envision that continued application of bioorthogonal chemistry, optogenetics, and directed evolution will yield new tools and discoveries to interrogate the phospholipidome and reveal new mechanisms regulating phospholipid homeostasis and roles for phospholipids in cell signaling.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





References
-
- Bumpus TW; Baskin JM Clickable Substrate Mimics Enable Imaging of Phospholipase D Activity. ACS Cent. Sci 2017, 3 (10), 1070–1077. - PMC - PubMed
-
This study showed that azido primary alcohols could replace water as a substrate of phospholipase D (PLD) enzymes, enabling the generation of fluorescent lipid reporters of PLD signaling in live cells in a method termed IMPACT (Imaging PLD Activity with Clickable Alcohols via Transphosphatidylation).
-
- Liang D; Wu K; Tei R; Bumpus TW; Ye J; Baskin JM A Real-Time, Click Chemistry Imaging Approach Reveals Stimulus-Specific Subcellular Locations of Phospholipase D Activity. Proc. Natl. Acad. Sci. U.S.A 2019, 116 (31), 15453–15462. - PMC - PubMed
-
This study developed a real-time variant of IMPACT harnessing a rapid and fluorogenic tetrazine ligation for imaging the subcellular localization of endogenous PLD signaling, revealing that such signaling via the protein kinase C–PLD pathway occurs at the plasma membrane and also visualizing rapid interorganelle bulk phospholipid transport from the plasma membrane to the endoplasmic reticulum on the second-to-minute time scale.
-
- Tei R; Baskin JM Spatiotemporal Control of Phosphatidic Acid Signaling with Optogenetic, Engineered Phospholipase Ds. J. Cell Biol 2020, 219 (3), No. e201907013. - PMC - PubMed
-
This study developed a light-controlled, optogenetic PLD that upon blue-light illumination could be recruited to target organelles to enable local synthesis of phosphatidic acid (PA) lipids and applied this tool to discover that pools of PA at the plasma membrane can selectively suppress oncogenic Hippo signaling.
-
- Bumpus TW; Huang S; Tei R; Baskin JM Click Chemistry-Enabled CRISPR Screening Reveals GSK3 as a Regulator of PLD Signaling. Proc. Natl. Acad. Sci. U.S.A 2021, 118 (48), No. e2025265118. - PMC - PubMed
-
This study combined IMPACT with pooled, genome-wide CRISPR interference screening to elucidate GSK3 as an activator of protein kinase C-stimulated PLD signaling, expanding the scope of unbiased CRISPR screening to the discovery of new regulators of enzyme-driven signaling pathways using bioorthogonal fluorescent labeling as a readout.
-
- Holthuis JCM; Menon AK Lipid Landscapes and Pipelines in Membrane Homeostasis. Nature 2014, 510 (7503), 48–57. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

