Shared cis-regulatory modules control expression of the tandem paralogs midline and H15 in the follicular epithelium
- PMID: 36278857
- PMCID: PMC9845738
- DOI: 10.1242/dev.201016
Shared cis-regulatory modules control expression of the tandem paralogs midline and H15 in the follicular epithelium
Abstract
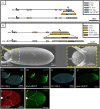
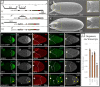
The posterior end of the follicular epithelium is patterned by midline (MID) and its paralog H15, the Drosophila homologs of the mammalian Tbx20 transcription factor. We have previously identified two cis-regulatory modules (CRMs) that recapitulate the endogenous pattern of mid in the follicular epithelium. Here, using CRISPR/Cas9 genome editing, we demonstrate redundant activity of these mid CRMs. Although the deletion of either CRM alone generated marginal change in mid expression, the deletion of both CRMs reduced expression by 60%. Unexpectedly, the deletion of the 5' proximal CRM of mid eliminated H15 expression. Interestingly, expression of these paralogs in other tissues remained unaffected in the CRM deletion backgrounds. These results suggest that the paralogs are regulated by a shared CRM that coordinates gene expression during posterior fate determination. The consistent overlapping expression of mid and H15 in various tissues may indicate that the paralogs could also be under shared regulation by other CRMs in these tissues.
Keywords: Cis-regulation; Anterior-posterior axis coordination; Developmental genes; Gene regulatory network; Genes tagging; Shared enhancers.
© 2022. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

