Molecular Diagnostic Yield of Exome Sequencing and Chromosomal Microarray in Cerebral Palsy: A Systematic Review and Meta-analysis
- PMID: 36279113
- PMCID: PMC9593320
- DOI: 10.1001/jamaneurol.2022.3549
Molecular Diagnostic Yield of Exome Sequencing and Chromosomal Microarray in Cerebral Palsy: A Systematic Review and Meta-analysis
Abstract
Importance: There are many known acquired risk factors for cerebral palsy (CP), but in some cases, CP is evident without risk factors (cryptogenic CP). Early CP cohort studies report a wide range of diagnostic yields for sequence variants assessed by exome sequencing (ES) and copy number variants (CNVs) assessed by chromosomal microarray (CMA).
Objective: To synthesize the emerging CP genetics literature and address the question of what percentage of individuals with CP have a genetic disorder via ES and CMA.
Data sources: Searched articles were indexed by PubMed with relevant queries pertaining to CP and ES/CMA (query date, March 15, 2022).
Study selection: Inclusion criteria were as follows: primary research study, case series with 10 or more nonrelated individuals, CP diagnosis, and ES and/or CMA data used for genetic evaluation. Nonblinded review was performed.
Data extraction and synthesis: Preferred Reporting Items for Systematic Reviews and Meta-analyses guidelines were used for assessing data quality and validity. Data were extracted by a single observer.
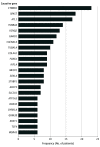
Main outcomes and measures: A separate meta-analysis was performed for each modality (ES, CMA). The primary outcome was proportion/molecular diagnostic yield (number of patients with a discovered genetic disorder divided by the total number of patients in the cohort), evaluated via meta-analysis of single proportions using random-effects logistic regression. A subgroup meta-analysis was conducted, using risk factor classification as a subgroup. A forest plot was used to display diagnostic yields of individual studies.
Results: In the meta-analysis of ES yield in CP, the overall diagnostic yield of ES among the cohorts (15 study cohorts comprising 2419 individuals from 11 articles) was 23% (95% CI, 15%-34%). The diagnostic yield across cryptogenic CP cohorts was 35% (95% CI, 27%-45%), compared with 7% (95% CI, 4%-12%) across cohorts with known risk factors (noncryptogenic CP). In the meta-analysis of CMA yield in CP, the diagnostic yield of CMA among the cohorts (5 study cohorts comprising 294 individuals from 5 articles) was 5% (95% CI, 2%-12%).
Conclusions and relevance: Results of this systematic review and meta-analysis suggest that for individuals with cryptogenic CP, ES followed by CMA to identify molecular disorders may be warranted.
Conflict of interest statement
Figures



References
-
- Rosenbaum P, Paneth N, Leviton A, et al. . A report: the definition and classification of cerebral palsy April 2006. Dev Med Child Neurol Suppl. 2007;109:8-14. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

