Phosphoinositide species and filamentous actin formation mediate engulfment by senescent tumor cells
- PMID: 36279312
- PMCID: PMC9632905
- DOI: 10.1371/journal.pbio.3001858
Phosphoinositide species and filamentous actin formation mediate engulfment by senescent tumor cells
Abstract
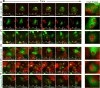
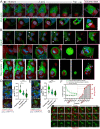
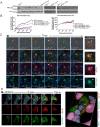
Cancer cells survive chemotherapy and cause lethal relapse by entering a senescent state that facilitates expression of many phagocytosis/macrophage-related genes that engender a novel cannibalism phenotype. We used biosensors and live-cell imaging to reveal the basic steps and mechanisms of engulfment by senescent human and mouse tumor cells. We show filamentous actin in predator cells was localized to the prey cell throughout the process of engulfment. Biosensors to various phosphoinositide (PI) species revealed increased concentration and distinct localization of predator PI(4) P and PI(4,5)P2 at the prey cell during early stages of engulfment, followed by a transient burst of PI(3) P before and following internalization. PIK3C2B, the kinase responsible for generating PI(3)P, was required for complete engulfment. Inhibition or knockdown of Clathrin, known to associate with PIK3C2B and PI(4,5)P2, severely impaired engulfment. In sum, our data reveal the most fundamental cellular processes of senescent cell engulfment, including the precise localizations and dynamics of actin and PI species throughout the entire process.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Bertheau P, Turpin E, Rickman DS, Espie M, de Reynies A, Feugeas JP, et al.. Exquisite sensitivity of TP53 mutant and basal breast cancers to a dose-dense epirubicin-cyclophosphamide regimen. PLoS Med. 2007;4(3):e90. Epub 2007/03/29. 06-PLME-RA-0470R3 [pii]. doi: 10.1371/journal.pmed.0040090 ; PubMed Central PMCID: PMC1831731. - DOI - PMC - PubMed
-
- Jackson JG, Pant V, Li Q, Chang LL, Quintas-Cardama A, Garza D, et al.. p53-Mediated Senescence Impairs the Apoptotic Response to Chemotherapy and Clinical Outcome in Breast Cancer. Cancer Cell. 2012;21(6):793–806. Epub 2012/06/16. doi: 10.1016/j.ccr.2012.04.027 ; PubMed Central PMCID: PMC3376352. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

