Ornithine decarboxylase supports ILC3 responses in infectious and autoimmune colitis through positive regulation of IL-22 transcription
- PMID: 36279426
- PMCID: PMC9659397
- DOI: 10.1073/pnas.2214900119
Ornithine decarboxylase supports ILC3 responses in infectious and autoimmune colitis through positive regulation of IL-22 transcription
Abstract
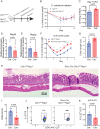
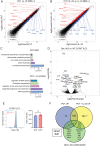
Group 3 innate lymphoid cells (ILC3s) are RORγT+ lymphocytes that are predominately enriched in mucosal tissues and produce IL-22 and IL-17A. They are the innate counterparts of Th17 cells. While Th17 lymphocytes utilize unique metabolic pathways in their differentiation program, it is unknown whether ILC3s make similar metabolic adaptations. We employed single-cell RNA sequencing and metabolomic profiling of intestinal ILC subsets to identify an enrichment of polyamine biosynthesis in ILC3s, converging on the rate-limiting enzyme ornithine decarboxylase (ODC1). In vitro and in vivo studies demonstrated that exogenous supplementation with the polyamine putrescine or its biosynthetic substrate, ornithine, enhanced ILC3 production of IL-22. Conditional deletion of ODC1 in ILC3s impaired mouse antibacterial defense against Citrobacter rodentium infection, which was associated with a decrease in anti-microbial peptide production by the intestinal epithelium. Furthermore, in a model of anti-CD40 colitis, deficiency of ODC1 in ILC3s markedly reduced the production of IL-22 and severity of inflammatory colitis. We conclude that ILC3-intrinsic polyamine biosynthesis facilitates efficient defense against enteric pathogens as well as exacerbates autoimmune colitis, thus representing an attractive target to modulate ILC3 function in intestinal disease.
Keywords: IL-22; enteritis; innate lymphoid cells; ornithine decarboxylase; polyamines.
Conflict of interest statement
M.C. receives research support from Pfizer. R.J.X. is a cofounder of Celsius Therapeutics. All the other authors have nothing to disclose.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

