Topological gelation of reconnecting polymers
- PMID: 36279471
- PMCID: PMC9636914
- DOI: 10.1073/pnas.2207728119
Topological gelation of reconnecting polymers
Abstract
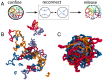
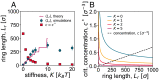
DNA recombination is a ubiquitous process that ensures genetic diversity. Contrary to textbook pictures, DNA recombination, as well as generic DNA translocations, occurs in a confined and highly entangled environment. Inspired by this observation, here, we investigate a solution of semiflexible polymer rings undergoing generic cutting and reconnection operations under spherical confinement. Our setup may be realized using engineered DNA in the presence of recombinase proteins or by considering micelle-like components able to form living (or reversibly breakable) polymer rings. We find that in such systems, there is a topological gelation transition, which can be triggered by increasing either the stiffness or the concentration of the rings. Flexible or dilute polymers break into an ensemble of short, unlinked, and segregated rings, whereas sufficiently stiff or dense polymers self-assemble into a network of long, linked, and mixed loops, many of which are knotted. We predict that the two phases should behave qualitatively differently in elution experiments monitoring the escape dynamics from a permeabilized container. Besides shedding some light on the biophysics and topology of genomes undergoing DNA reconnection in vivo, our findings could be leveraged in vitro to design polymeric complex fluids-e.g., DNA-based complex fluids or living polymer networks-with desired topologies.
Keywords: DNA topology; MD simulations; living polymers; topological gel.
Conflict of interest statement
The authors declare no competing interest.
Figures



References
-
- Alberts B., Johnson A., Lewis J., Morgan D., Raff M., Molecular Biology of the Cell (Taylor & Francis, Abingdon, UK, 2014).
-
- Hallet B., Sherratt D. J., Transposition and site-specific recombination: Adapting DNA cut-and-paste mechanisms to a variety of genetic rearrangements. FEMS Microbiol. Rev. 21, 157–178 (1997). - PubMed
-
- Pathania S., Jayaram M., Harshey R. M., Path of DNA within the Mu transpososome. Transposase interactions bridging two Mu ends and the enhancer trap five DNA supercoils. Cell 109, 425–436 (2002). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

