A peptide array pipeline for the development of Spike-ACE2 interaction inhibitors
- PMID: 36279985
- PMCID: PMC9585897
- DOI: 10.1016/j.peptides.2022.170898
A peptide array pipeline for the development of Spike-ACE2 interaction inhibitors
Abstract
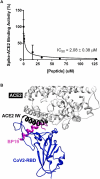
In humans, coronaviruses are the cause of endemic illness and have been the causative agents of more severe epidemics. Most recently, SARS-CoV-2 was the causative agent of the COVID19 pandemic. Thus, there is a high interest in developing therapeutic agents targeting various stages of the coronavirus viral life cycle to disrupt viral propagation. Besides the development of small-molecule therapeutics that target viral proteases, there is also interest molecular tools to inhibit the initial event of viral attachment of the SARS-CoV-2 Spike protein to host ACE2 surface receptor. Here, we leveraged known structural information and peptide arrays to develop an in vitro peptide inhibitor of the Spike-ACE2 interaction. First, from previous co-crystal structures of the Spike-ACE2 complex, we identified an initial 24-residue long region (sequence: STIEEQAKTFLDKFNHEAEDLFYQ) on the ACE2 sequence that encompasses most of the known contact residues. Next, we scanned this 24-mer window along the ACE2 N-terminal helix and found that maximal binding to the SARS-CoV-2 receptor binding domain (CoV2-RBD) was increased when this window was shifted nine residues in the N-terminal direction. Further, by systematic permutation of this shifted ACE2-derived peptide we identified mutations to the wildtype sequence that confer increased binding of the CoV2-RBD. Among these peptides, we identified binding peptide 19 (referred to as BP19; sequence: SLVAVTAAQSTIEEQAKTFLDKFI) as an in vitro inhibitor of the Spike-ACE2 interaction with an IC50 of 2.08 ± 0.38 μM. Overall, BP19 adds to the arsenal of Spike-ACE2 inhibitors, and this study highlights the utility of systematic peptide arrays as a platform for the development of coronavirus protein inhibitors.
Keywords: ACE2; Peptide array; Peptide inhibitor; Spike.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare no conflict of interest.
Figures

Similar articles
-
An angiotensin-converting enzyme-2-derived heptapeptide GK-7 for SARS-CoV-2 spike blockade.Peptides. 2021 Nov;145:170638. doi: 10.1016/j.peptides.2021.170638. Epub 2021 Aug 19. Peptides. 2021. PMID: 34419496 Free PMC article.
-
A Collection of Designed Peptides to Target SARS-CoV-2 Spike RBD-ACE2 Interaction.Int J Mol Sci. 2021 Oct 27;22(21):11627. doi: 10.3390/ijms222111627. Int J Mol Sci. 2021. PMID: 34769056 Free PMC article.
-
Stapled Peptides Based on Human Angiotensin-Converting Enzyme 2 (ACE2) Potently Inhibit SARS-CoV-2 Infection In Vitro.mBio. 2020 Dec 11;11(6):e02451-20. doi: 10.1128/mBio.02451-20. mBio. 2020. PMID: 33310780 Free PMC article.
-
Inhibition of S-protein RBD and hACE2 Interaction for Control of SARSCoV- 2 Infection (COVID-19).Mini Rev Med Chem. 2021;21(6):689-703. doi: 10.2174/1389557520666201117111259. Mini Rev Med Chem. 2021. PMID: 33208074 Review.
-
Interactions of angiotensin-converting enzyme-2 (ACE2) and SARS-CoV-2 spike receptor-binding domain (RBD): a structural perspective.Mol Biol Rep. 2023 Mar;50(3):2713-2721. doi: 10.1007/s11033-022-08193-4. Epub 2022 Dec 23. Mol Biol Rep. 2023. PMID: 36562937 Free PMC article. Review.
Cited by
-
Investigating In Situ Expression of c-MYC and Candidate Ubiquitin-Specific Proteases in DLBCL and Assessment for Peptidyl Disruptor Molecule against c-MYC-USP37 Complex.Molecules. 2023 Mar 7;28(6):2441. doi: 10.3390/molecules28062441. Molecules. 2023. PMID: 36985413 Free PMC article.
-
Progress in SARS-CoV-2, diagnostic and clinical treatment of COVID-19.Heliyon. 2024 Jun 17;10(12):e33179. doi: 10.1016/j.heliyon.2024.e33179. eCollection 2024 Jun 30. Heliyon. 2024. PMID: 39021908 Free PMC article. Review.
-
Peptide-Based Inhibitors of Protein-Protein Interactions (PPIs): A Case Study on the Interaction Between SARS-CoV-2 Spike Protein and Human Angiotensin-Converting Enzyme 2 (hACE2).Biomedicines. 2024 Oct 16;12(10):2361. doi: 10.3390/biomedicines12102361. Biomedicines. 2024. PMID: 39457672 Free PMC article. Review.
References
-
- Zhong N.S., Zheng B.J., Li Y.M., Poon L.L.M., Xie Z.H., Chan K.H., Li P.H., Tan S.Y., Chang Q., Xie J.P., et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet. 2003;362:1353. doi: 10.1016/S0140-6736(03)14630-2. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

