Ribosome-mediated biosynthesis of pyridazinone oligomers in vitro
- PMID: 36280685
- PMCID: PMC9592601
- DOI: 10.1038/s41467-022-33701-2
Ribosome-mediated biosynthesis of pyridazinone oligomers in vitro
Abstract
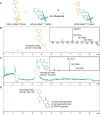
The ribosome is a macromolecular machine that catalyzes the sequence-defined polymerization of L-α-amino acids into polypeptides. The catalysis of peptide bond formation between amino acid substrates is based on entropy trapping, wherein the adjacency of transfer RNA (tRNA)-coupled acyl bonds in the P-site and the α-amino groups in the A-site aligns the substrates for coupling. The plasticity of this catalytic mechanism has been observed in both remnants of the evolution of the genetic code and modern efforts to reprogram the genetic code (e.g., ribosomal incorporation of non-canonical amino acids, ribosomal ester formation). However, the limits of ribosome-mediated polymerization are underexplored. Here, rather than peptide bonds, we demonstrate ribosome-mediated polymerization of pyridazinone bonds via a cyclocondensation reaction between activated γ-keto and α-hydrazino ester monomers. In addition, we demonstrate the ribosome-catalyzed synthesis of peptide-hybrid oligomers composed of multiple sequence-defined alternating pyridazinone linkages. Our results highlight the plasticity of the ribosome's ancient bond-formation mechanism, expand the range of non-canonical polymeric backbones that can be synthesized by the ribosome, and open the door to new applications in synthetic biology.
© 2022. The Author(s).
Conflict of interest statement
M.C.J., E.V.A., J.Lee, J.Lim, and J.N.C. are co-inventors on the US provisional patent application that incorporates discoveries described in this manuscript (US Patent Application Serial No. 63/144,814). M.C.J. has a financial interest in Pearl Bio. M.C.J.’s interests are reviewed and managed by Northwestern University in accordance with their conflict-of-interest policies. The remaining authors declare no competing interests.
Figures



Similar articles
-
Expanding the limits of the second genetic code with ribozymes.Nat Commun. 2019 Nov 8;10(1):5097. doi: 10.1038/s41467-019-12916-w. Nat Commun. 2019. PMID: 31704912 Free PMC article.
-
Expanded ribosomal synthesis of non-standard cyclic backbones in vitro.Nat Commun. 2025 May 28;16(1):4957. doi: 10.1038/s41467-025-60126-4. Nat Commun. 2025. PMID: 40436849 Free PMC article.
-
Ribosome-mediated polymerization of long chain carbon and cyclic amino acids into peptides in vitro.Nat Commun. 2020 Aug 27;11(1):4304. doi: 10.1038/s41467-020-18001-x. Nat Commun. 2020. PMID: 32855412 Free PMC article.
-
Mechanism of peptide bond formation on the ribosome.Q Rev Biophys. 2006 Aug;39(3):203-25. doi: 10.1017/S003358350600429X. Epub 2006 Aug 8. Q Rev Biophys. 2006. PMID: 16893477 Review.
-
D Amino Acids Highlight the Catalytic Power of the Ribosome.Cell Chem Biol. 2019 Dec 19;26(12):1639-1641. doi: 10.1016/j.chembiol.2019.10.007. Epub 2019 Oct 31. Cell Chem Biol. 2019. PMID: 31680066 Review.
Cited by
-
Engineering tRNAs for the Ribosomal Translation of Non-proteinogenic Monomers.Chem Rev. 2024 May 22;124(10):6444-6500. doi: 10.1021/acs.chemrev.3c00894. Epub 2024 Apr 30. Chem Rev. 2024. PMID: 38688034 Free PMC article. Review.
-
Tuning tRNAs for improved translation.Front Genet. 2024 Jun 25;15:1436860. doi: 10.3389/fgene.2024.1436860. eCollection 2024. Front Genet. 2024. PMID: 38983271 Free PMC article. Review.
-
A Translation-Independent Directed Evolution Strategy to Engineer Aminoacyl-tRNA Synthetases.ACS Cent Sci. 2024 May 20;10(6):1211-1220. doi: 10.1021/acscentsci.3c01557. eCollection 2024 Jun 26. ACS Cent Sci. 2024. PMID: 38947215 Free PMC article.
-
Expanding the substrate scope of pyrrolysyl-transfer RNA synthetase enzymes to include non-α-amino acids in vitro and in vivo.Nat Chem. 2023 Jul;15(7):960-971. doi: 10.1038/s41557-023-01224-y. Epub 2023 Jun 1. Nat Chem. 2023. PMID: 37264106 Free PMC article.
-
Direct and quantitative analysis of tRNA acylation using intact tRNA liquid chromatography-mass spectrometry.Nat Protoc. 2025 May;20(5):1246-1274. doi: 10.1038/s41596-024-01086-9. Epub 2025 Jan 6. Nat Protoc. 2025. PMID: 39762443 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

