Advances in CRISPR therapeutics
- PMID: 36280707
- PMCID: PMC9589773
- DOI: 10.1038/s41581-022-00636-2
Advances in CRISPR therapeutics
Abstract
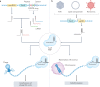
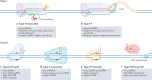
The clustered regularly interspaced short palindromic repeats (CRISPR) renaissance was catalysed by the discovery that RNA-guided prokaryotic CRISPR-associated (Cas) proteins can create targeted double-strand breaks in mammalian genomes. This finding led to the development of CRISPR systems that harness natural DNA repair mechanisms to repair deficient genes more easily and precisely than ever before. CRISPR has been used to knock out harmful mutant genes and to fix errors in coding sequences to rescue disease phenotypes in preclinical studies and in several clinical trials. However, most genetic disorders result from combinations of mutations, deletions and duplications in the coding and non-coding regions of the genome and therefore require sophisticated genome engineering strategies beyond simple gene knockout. To overcome this limitation, the toolbox of natural and engineered CRISPR-Cas systems has been dramatically expanded to include diverse tools that function in human cells for precise genome editing and epigenome engineering. The application of CRISPR technology to edit the non-coding genome, modulate gene regulation, make precise genetic changes and target infectious diseases has the potential to lead to curative therapies for many previously untreatable diseases.
© 2022. Springer Nature Limited.
Conflict of interest statement
L.S.Q. is a founder and scientific adviser of Epic Bio, and a scientific adviser of Laboratory of Genomics Research (LGR). The other authors declare no competing interests.
Figures


References
-
- Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987;169:5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

