Resolution doubling in light-sheet microscopy via oblique plane structured illumination
- PMID: 36280718
- PMCID: PMC10182454
- DOI: 10.1038/s41592-022-01635-8
Resolution doubling in light-sheet microscopy via oblique plane structured illumination
Abstract
Structured illumination microscopy (SIM) doubles the spatial resolution of a fluorescence microscope without requiring high laser powers or specialized fluorophores. However, the excitation of out-of-focus fluorescence can accelerate photobleaching and phototoxicity. In contrast, light-sheet fluorescence microscopy (LSFM) largely avoids exciting out-of-focus fluorescence, thereby enabling volumetric imaging with low photobleaching and intrinsic optical sectioning. Combining SIM with LSFM would enable gentle three-dimensional (3D) imaging at doubled resolution. However, multiple orientations of the illumination pattern, which are needed for isotropic resolution doubling in SIM, are challenging to implement in a light-sheet format. Here we show that multidirectional structured illumination can be implemented in oblique plane microscopy, an LSFM technique that uses a single objective for excitation and detection, in a straightforward manner. We demonstrate isotropic lateral resolution below 150 nm, combined with lower phototoxicity compared to traditional SIM systems and volumetric acquisition speed exceeding 1 Hz.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing Interests
R.F., B.C. and B.J.C. have filed a patent for the image rotator and applications to microscopy.
Figures

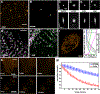
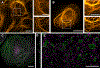
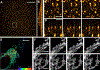
References
-
- Liu Z, Lavis, Luke D & Betzig E Imaging Live-Cell Dynamics and Structure at the Single-Molecule Level. Molecular Cell 58, 644–659 (2015). - PubMed
-
- Stelzer EHK et al. Light sheet fluorescence microscopy. Nature Reviews Methods Primers 1, 73 (2021).
-
- Olarte OE, Andilla J, Gualda EJ & Loza-Alvarez P Light-sheet microscopy: a tutorial. Advances in Optics and Photonics 10 (2018).
-
- Stelzer EHK Light-sheet fluorescence microscopy for quantitative biology. Nature Methods 12, 23–26 (2014). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

