Analyses of the autism-associated neuroligin-3 R451C mutation in human neurons reveal a gain-of-function synaptic mechanism
- PMID: 36280753
- PMCID: PMC10123180
- DOI: 10.1038/s41380-022-01834-x
Analyses of the autism-associated neuroligin-3 R451C mutation in human neurons reveal a gain-of-function synaptic mechanism
Abstract
Mutations in many synaptic genes are associated with autism spectrum disorders (ASD), suggesting that synaptic dysfunction is a key driver of ASD pathogenesis. Among these mutations, the R451C substitution in the NLGN3 gene that encodes the postsynaptic adhesion molecule Neuroligin-3 is noteworthy because it was the first specific mutation linked to ASDs. In mice, the corresponding Nlgn3 R451C-knockin mutation recapitulates social interaction deficits of ASD patients and produces synaptic abnormalities, but the impact of the NLGN3 R451C mutation on human neurons has not been investigated. Here, we generated human knockin neurons with the NLGN3 R451C and NLGN3 null mutations. Strikingly, analyses of NLGN3 R451C-mutant neurons revealed that the R451C mutation decreased NLGN3 protein levels but enhanced the strength of excitatory synapses without affecting inhibitory synapses; meanwhile NLGN3 knockout neurons showed reduction in excitatory synaptic strengths. Moreover, overexpression of NLGN3 R451C recapitulated the synaptic enhancement in human neurons. Notably, the augmentation of excitatory transmission was confirmed in vivo with human neurons transplanted into mouse forebrain. Using single-cell RNA-seq experiments with co-cultured excitatory and inhibitory NLGN3 R451C-mutant neurons, we identified differentially expressed genes in relatively mature human neurons corresponding to synaptic gene expression networks. Moreover, gene ontology and enrichment analyses revealed convergent gene networks associated with ASDs and other mental disorders. Our findings suggest that the NLGN3 R451C mutation induces a gain-of-function enhancement in excitatory synaptic transmission that may contribute to the pathophysiology of ASD.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Figures
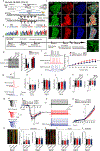

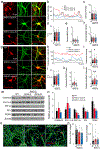
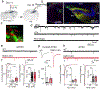

References
-
- Skuse DH, Mandy WP, Scourfield J. Measuring autistic traits: heritability, reliability and validity of the Social and Communication Disorders Checklist. Br J Psychiatry. 2005;187:568–72. - PubMed
MeSH terms
Substances
Grants and funding
- F30 MH108321/MH/NIMH NIH HHS/United States
- R01 NS102382/NS/NINDS NIH HHS/United States
- R01 MH092931/MH/NIMH NIH HHS/United States
- R01 MH122519/MH/NIMH NIH HHS/United States
- R21 MH126420/MH/NIMH NIH HHS/United States
- MH126420/U.S. Department of Health & Human Services | NIH | National Institute of Mental Health (NIMH)
- F30MH108321/U.S. Department of Health & Human Services | NIH | National Institute of Mental Health (NIMH)
- MH122519/U.S. Department of Health & Human Services | NIH | National Institute of Mental Health (NIMH)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

