Heterologously secreted MbxA from Moraxella bovis induces a membrane blebbing response of the human host cell
- PMID: 36280777
- PMCID: PMC9592583
- DOI: 10.1038/s41598-022-22480-x
Heterologously secreted MbxA from Moraxella bovis induces a membrane blebbing response of the human host cell
Abstract
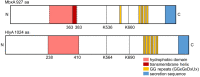
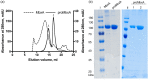
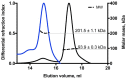
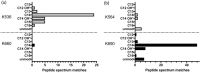
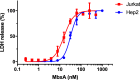
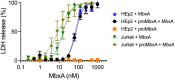
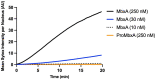
Many proteins of the Repeats in Toxins (RTX) protein family are toxins of Gram-negative pathogens including hemolysin A (HlyA) of uropathogenic E. coli. RTX proteins are secreted via Type I secretion systems (T1SS) and adopt their native conformation in the Ca2+-rich extracellular environment. Here we employed the E. coli HlyA T1SS as a heterologous surrogate system for the RTX toxin MbxA from the bovine pathogen Moraxella bovis. In E. coli the HlyA system successfully activates the heterologous MbxA substrate by acylation and secretes the precursor proMbxA and active MbxA allowing purification of both species in quantities sufficient for a variety of investigations. The activating E. coli acyltransferase HlyC recognizes the acylation sites in MbxA, but unexpectedly in a different acylation pattern as for its endogenous substrate HlyA. HlyC-activated MbxA shows host species-independent activity including a so-far unknown toxicity against human lymphocytes and epithelial cells. Using live-cell imaging, we show an immediate MbxA-mediated permeabilization and a rapidly developing blebbing of the plasma membrane in epithelial cells, which is associated with immediate cell death.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Escherichia coli hemolysin mutants with altered target cell specificity.Infect Immun. 1996 Aug;64(8):3081-7. doi: 10.1128/iai.64.8.3081-3087.1996. Infect Immun. 1996. PMID: 8757837 Free PMC article.
-
Quantification and Surface Localization of the Hemolysin A Type I Secretion System at the Endogenous Level and under Conditions of Overexpression.Appl Environ Microbiol. 2022 Feb 8;88(3):e0189621. doi: 10.1128/AEM.01896-21. Epub 2021 Dec 1. Appl Environ Microbiol. 2022. PMID: 34851699 Free PMC article.
-
In vivo proteolytic degradation of the Escherichia coli acyltransferase HlyC.J Biol Chem. 2001 May 18;276(20):16660-6. doi: 10.1074/jbc.M009514200. Epub 2001 Feb 15. J Biol Chem. 2001. PMID: 11278516
-
Acylation of Escherichia coli hemolysin: a unique protein lipidation mechanism underlying toxin function.Microbiol Mol Biol Rev. 1998 Jun;62(2):309-33. doi: 10.1128/MMBR.62.2.309-333.1998. Microbiol Mol Biol Rev. 1998. PMID: 9618444 Free PMC article. Review.
-
Redefining the bacterial Type I protein secretion system.Adv Microb Physiol. 2023;82:155-204. doi: 10.1016/bs.ampbs.2022.10.003. Epub 2022 Dec 5. Adv Microb Physiol. 2023. PMID: 36948654 Review.
Cited by
-
Type 1 secretion necessitates a tight interplay between all domains of the ABC transporter.Sci Rep. 2024 Apr 18;14(1):8994. doi: 10.1038/s41598-024-59759-0. Sci Rep. 2024. PMID: 38637678 Free PMC article.
-
Investigations on the substrate binding sites of hemolysin B, an ABC transporter, of a type 1 secretion system.Front Microbiol. 2022 Dec 1;13:1055032. doi: 10.3389/fmicb.2022.1055032. eCollection 2022. Front Microbiol. 2022. PMID: 36532430 Free PMC article.
-
Moraxella haemolytica sp. nov., isolated from a goat with respiratory disease.Arch Microbiol. 2023 Dec 28;206(1):45. doi: 10.1007/s00203-023-03782-8. Arch Microbiol. 2023. PMID: 38153526
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

