CCHCR1-astrin interaction promotes centriole duplication through recruitment of CEP72
- PMID: 36280838
- PMCID: PMC9590400
- DOI: 10.1186/s12915-022-01437-6
CCHCR1-astrin interaction promotes centriole duplication through recruitment of CEP72
Abstract
Background: The centrosome is one of the most important non-membranous organelles regulating microtubule organization and progression of cell mitosis. The coiled-coil alpha-helical rod protein 1 (CCHCR1, also known as HCR) gene is considered to be a psoriasis susceptibility gene, and the protein is suggested to be localized to the P-bodies and centrosomes in mammalian cells. However, the exact cellular function of HCR and its potential regulatory role in the centrosomes remain unexplored.
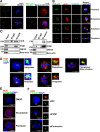
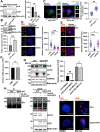
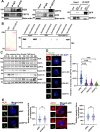
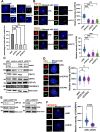
Results: We found that HCR interacts directly with astrin, a key factor in centrosome maturation and mitosis. Immunoprecipitation assays showed that the coiled-coil region present in the C-terminus of HCR and astrin respectively mediated the interaction between them. Astrin not only recruits HCR to the centrosome, but also protects HCR from ubiquitin-proteasome-mediated degradation. In addition, depletion of either HCR or astrin significantly reduced centrosome localization of CEP72 and subsequent MCPH proteins, including CEP152, CDK5RAP2, and CEP63. The absence of HCR also caused centriole duplication defects and mitotic errors, resulting in multipolar spindle formation, genomic instability, and DNA damage.
Conclusion: We conclude that HCR is localized and stabilized at the centrosome by directly binding to astrin. HCR are required for the centrosomal recruitment of MCPH proteins and centriolar duplication. Both HCR and astrin play key roles in keeping normal microtubule assembly and maintaining genomic stability.
Keywords: Astrin; CCHCR1; CEP72; Centrosome; Microtubule organization; Mitosis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. - PubMed
-
- Hixon ML, Gualberto A. The control of mitosis. Front Biosci. 2000;5:D50–D57. - PubMed
-
- Musacchio A, Hardwick KG. The spindle checkpoint: structural insights into dynamic signalling. Nat Rev Mol Cell Biol. 2002;3:731–741. - PubMed
-
- Nunes V, Ferreira JG. From the cytoskeleton to the nucleus: an integrated view on early spindle assembly. Semin Cell Dev Biol. 2021. 10.1016/j.semcdb.2021.03.006. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

