A machine learning study of COVID-19 serology and molecular tests and predictions
- PMID: 36281350
- PMCID: PMC9583626
- DOI: 10.1016/j.smhl.2022.100331
A machine learning study of COVID-19 serology and molecular tests and predictions
Abstract
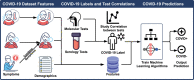
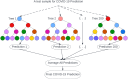
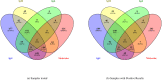
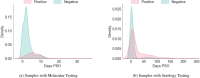
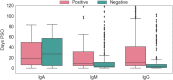
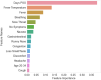
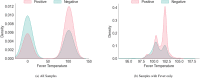
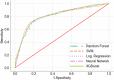
Serology and molecular tests are the two most commonly used methods for rapid COVID-19 infection testing. The two types of tests have different mechanisms to detect infection, by measuring the presence of viral SARS-CoV-2 RNA (molecular test) or detecting the presence of antibodies triggered by the SARS-CoV-2 virus (serology test). A handful of studies have shown that symptoms, combined with demographic and/or diagnosis features, can be helpful for the prediction of COVID-19 test outcomes. However, due to nature of the test, serology and molecular tests vary significantly. There is no existing study on the correlation between serology and molecular tests, and what type of symptoms are the key factors indicating the COVID-19 positive tests. In this study, we propose a machine learning based approach to study serology and molecular tests, and use features to predict test outcomes. A total of 2,467 donors, each tested using one or multiple types of COVID-19 tests, are collected as our testbed. By cross checking test types and results, we study correlation between serology and molecular tests. For test outcome prediction, we label 2,467 donors as positive or negative, by using their serology or molecular test results, and create symptom features to represent each donor for learning. Because COVID-19 produces a wide range of symptoms and the data collection process is essentially error prone, we group similar symptoms into bins. This decreases the feature space and sparsity. Using binned symptoms, combined with demographic features, we train five classification algorithms to predict COVID-19 test results. Experiments show that XGBoost achieves the best performance with 76.85% accuracy and 81.4% AUC scores, demonstrating that symptoms are indeed helpful for predicting COVID-19 test outcomes. Our study investigates the relationship between serology and molecular tests, identifies meaningful symptom features associated with COVID-19 infection, and also provides a way for rapid screening and cost effective detection of COVID-19 infection.
Keywords: 68T05; 68T50; 92C50; 92C55; 92C60; COVID-19; Classification; Machine Learning; Molecular test; Serology test; Symptoms.
© 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



Similar articles
-
Effectiveness and cost-effectiveness of four different strategies for SARS-CoV-2 surveillance in the general population (CoV-Surv Study): a structured summary of a study protocol for a cluster-randomised, two-factorial controlled trial.Trials. 2021 Jan 8;22(1):39. doi: 10.1186/s13063-020-04982-z. Trials. 2021. PMID: 33419461 Free PMC article.
-
The Development and Validation of Simplified Machine Learning Algorithms to Predict Prognosis of Hospitalized Patients With COVID-19: Multicenter, Retrospective Study.J Med Internet Res. 2022 Jan 21;24(1):e31549. doi: 10.2196/31549. J Med Internet Res. 2022. PMID: 34951865 Free PMC article.
-
Prevalence of Sars-Cov-2 Infection in Health Workers (HWs) and Diagnostic Test Performance: The Experience of a Teaching Hospital in Central Italy.Int J Environ Res Public Health. 2020 Jun 19;17(12):4417. doi: 10.3390/ijerph17124417. Int J Environ Res Public Health. 2020. PMID: 32575505 Free PMC article.
-
SARS-CoV-2 serology increases diagnostic accuracy in CT-suspected, PCR-negative COVID-19 patients during pandemic.Respir Res. 2021 Apr 23;22(1):119. doi: 10.1186/s12931-021-01717-9. Respir Res. 2021. PMID: 33892720 Free PMC article.
-
Effectiveness of tests to detect the presence of SARS-CoV-2 virus, and antibodies to SARS-CoV-2, to inform COVID-19 diagnosis: a rapid systematic review.BMJ Evid Based Med. 2022 Feb;27(1):33-45. doi: 10.1136/bmjebm-2020-111511. Epub 2020 Oct 1. BMJ Evid Based Med. 2022. PMID: 33004426
References
-
- Bishop C.M. 781058134; Springer: 2009. Pattern recognition and machine learning. OCLC.
-
- Chen, T., & Guestrin, C. (2016). XGBoost: A Scalable Tree Boosting System. In Proc. of the 22nd ACM SIGKDD Conf. (pp. 785–794). New York, NY, USA: ISBN: 9781450342322, 10.1145/2939672.2939785. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous
