A Multiplex Gene Expression Assay for Direct Measurement of RNA Transcripts in Crude Lysates of the Nematode Caenorhabditis elegans Used as a Bioanalytical Tool
- PMID: 36282018
- PMCID: PMC10107722
- DOI: 10.1002/etc.5505
A Multiplex Gene Expression Assay for Direct Measurement of RNA Transcripts in Crude Lysates of the Nematode Caenorhabditis elegans Used as a Bioanalytical Tool
Abstract
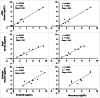
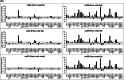
Gene expression profiling in Caenorhabditis elegans has been demonstrated to be a potential bioanalytical tool to detect the toxic potency of environmental contaminants. The RNA transcripts of genes responding to toxic exposure can be used as biomarkers for detecting these toxins. For routine application in environmental quality monitoring, an easy-to-use multiplex assay is required to reliably quantify expression levels of these biomarkers. In the present study, a bead-based assay was developed to fingerprint gene expression in C. elegans by quantitating messenger RNAs (mRNAs) of multiple target genes directly from crude nematode lysates, circumventing RNA extraction and purification steps. The assay uses signal amplification rather than target amplification for direct measurement of toxin-induced RNA transcripts. Using a 50-gene panel, the expression changes of four candidate reference genes and 46 target mRNAs for various contaminants and wastewaters were successfully measured, and the expression profiles indicated the type of toxin present. Moreover, the multiplex assay response was in line with previous results obtained with more time-consuming reverse-transcription quantitative polymerase chain reaction and microarray analyses. In addition, the transcriptomic profiles of nematodes exposed to wastewater samples and extracts prepared from tissues of swimming crabs were evaluated. The profiles indicated the presence of organic pollutants. The present study illustrates the successful development of a multiplex fluorescent bead-based approach using nematode C. elegans crude lysates for gene expression profiling of target RNAs. This method can be used to routinely fingerprint the presence of toxic contaminants in environmental samples and to identify the most biologically active fraction of the contaminant mixture in a toxicity identification and evaluation approach. Environ Toxicol Chem 2023;42:130-142. © 2022 The Authors. Environmental Toxicology and Chemistry published by Wiley Periodicals LLC on behalf of SETAC.
Keywords: Caenorhabditis elegans; biomarkers; multiplex assay; nematode bioassay; transcriptomics; water quality.
© 2022 The Authors. Environmental Toxicology and Chemistry published by Wiley Periodicals LLC on behalf of SETAC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Development of a transcription-based bioanalytical tool to quantify the toxic potencies of hydrophilic compounds in water using the nematode Caenorhabditis elegans.Ecotoxicol Environ Saf. 2021 Dec 20;227:112923. doi: 10.1016/j.ecoenv.2021.112923. Epub 2021 Oct 23. Ecotoxicol Environ Saf. 2021. PMID: 34700171
-
Determining Toxic Potencies of Water-Soluble Contaminants in Wastewater Influents and Effluent Using Gene Expression Profiling in C. elegans as a Bioanalytical Tool.Arch Environ Contam Toxicol. 2022 Oct;83(3):284-294. doi: 10.1007/s00244-022-00959-y. Epub 2022 Oct 3. Arch Environ Contam Toxicol. 2022. PMID: 36190544 Free PMC article.
-
Toxic effects of di(2-ethylhexyl)phthalate on mortality, growth, reproduction and stress-related gene expression in the soil nematode Caenorhabditis elegans.Toxicology. 2007 Jul 31;237(1-3):126-133. doi: 10.1016/j.tox.2007.05.008. Epub 2007 May 18. Toxicology. 2007. PMID: 17604895
-
Caenorhabditis elegans, a Biological Model for Research in Toxicology.Rev Environ Contam Toxicol. 2016;237:1-35. doi: 10.1007/978-3-319-23573-8_1. Rev Environ Contam Toxicol. 2016. PMID: 26613986 Review.
-
Functional genomic approaches using the nematode Caenorhabditis elegans as a model system.J Biochem Mol Biol. 2004 Jan 31;37(1):107-13. doi: 10.5483/bmbrep.2004.37.1.107. J Biochem Mol Biol. 2004. PMID: 14761308 Review.
Cited by
-
Unearthing roundworms: Nematodes as determinants of human health.One Health. 2025 Jun 13;21:101103. doi: 10.1016/j.onehlt.2025.101103. eCollection 2025 Dec. One Health. 2025. PMID: 40599644 Free PMC article. Review.
References
-
- Baugh, L. R. , Wen, J. C. , Hill, A. A. , Slonim, D. K. , Brown, E. L. , & Hunter, C. P. (2005). Synthetic lethal analysis of Caenorhabditis elegans posterior embryonic patterning genes identifies conserved genetic interactions. Genome Biology, 6(5), Article R45. 10.1186/gb-2005-6-5-r45 - DOI - PMC - PubMed
-
- Bodero, M. , Bovee, T. F. H. , Portier, L. , & Hendriksen, P. J. M. (2019). Detection and profiling of diarrheic marine biotoxins in shellfish by mRNA analysis of exposed Caco‐2 cells using qRT‐PCR and multiplex magnetic bead‐based assays. ALTEX: Alternatives to Animal Experimentation, 36(2), 203–214. 10.14573/altex.1805291 - DOI - PubMed
-
- Bourdon‐Lacombe, J. A. , Moffat, I. D. , Deveau, M. , Husain, M. , Auerbach, S. , Krewski, D. , Thomas, R. S. , Bushel, P. R. , Williams, A. , & Yauk, C. L. (2015). Technical guide for applications of gene expression profiling in human health risk assessment of environmental chemicals. Regulatory Toxicology and Pharmacology, 72(2), 292–309. 10.1016/j.yrtph.2015.04.010 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

